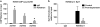Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2 - PubMed (original) (raw)
. 2017 Dec;22(12):1746-1758.
doi: 10.1038/mp.2016.131. Epub 2016 Aug 30.
A L Johnstone 2 3, B B Khomtchouk 2 3, J D Tapocik 4, C Pitcairn 4, F Rehman 4, E Augier 1, A Borich 4, J R Schank 5, C A Rienas 2 3, D J Van Booven 6, H Sun 4, D Nätt 1, C Wahlestedt 2 3, M Heilig 1 4
Affiliations
- PMID: 27573876
- PMCID: PMC5677579
- DOI: 10.1038/mp.2016.131
Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2
E Barbier et al. Mol Psychiatry. 2017 Dec.
Abstract
Epigenetic processes have been implicated in the pathophysiology of alcohol dependence, but the specific molecular mechanisms mediating dependence-induced neuroadaptations remain largely unknown. Here, we found that a history of alcohol dependence persistently decreased the expression of Prdm2, a histone methyltransferase that monomethylates histone 3 at the lysine 9 residue (H3K9me1), in the rat dorsomedial prefrontal cortex (dmPFC). Downregulation of Prdm2 was associated with decreased H3K9me1, supporting that changes in Prdm2 mRNA levels affected its activity. Chromatin immunoprecipitation followed by massively parallel DNA sequencing showed that genes involved in synaptic communication are epigenetically regulated by H3K9me1 in dependent rats. In non-dependent rats, viral-vector-mediated knockdown of Prdm2 in the dmPFC resulted in expression changes similar to those observed following a history of alcohol dependence. Prdm2 knockdown resulted in increased alcohol self-administration, increased aversion-resistant alcohol intake and enhanced stress-induced relapse to alcohol seeking, a phenocopy of postdependent rats. Collectively, these results identify a novel epigenetic mechanism that contributes to the development of alcohol-seeking behavior following a history of dependence.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare that, except for income received from our primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service, and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
Figures
Figure 1
(a) Validation of the histone 3 at the lysine 9 residue (H3K9me1) antibody. (b) Validation of the bioinformatic pipeline using quantitative real-time PCR (qPCR) to verify differential H3K9me1 enrichment at the Syt1 gene in control versus postdependent rats. *P < 0.05.
Figure 2
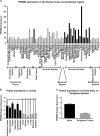
Prdm2 expression in the brain and peripheral organs. (a) Human Affymetrix microarray data (
). (b and c) Quantitative real-time PCR (qPCR) of adult mouse tissues. The central nervous system tissue is shown in black bars and the peripheral tissues are shown in gray bars. *P < 0.05.
Figure 3
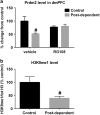
(a) Repeated cycles of alcohol intoxication decreased mRNA levels of Prdm2. The DNA methyltransferase (DNMT) inhibitor RG-108 significantly restored levels of Prdm2 (n = 6–10;two-way analysis of variance (ANOVA): main effect of group;control versus postdependent;F(1–29) = 6.3; P = 0.02; post hoc analysis control vehicle versus postdependent vehicle; P = 0.008; No significant difference between postdependent RG-108 versus control RG-108 or postdependent RG-108 versus control vehicle). The mean values (± s.e.m.) of control rats are shown in black bars. The mean values (± s.e.m.) of postdependent rats are shown in gray bars. mRNA expression is presented as % change from control/vehicle values. (b) Western blot analysis shows that monomethylation at H3K9 (H3K9me1) is significantly reduced in the dorsomedial prefrontal cortex (dmPFC) of postdependent rats (n = 7 per group;one-way ANOVA;main effect of group;control versus postdependent; F(1–12) = 7.1; P < 0.05). #P < 0.05, control versus postdependent rats.
Figure 4
(a–c) Expression of PRDM2 in rat dorsomedial prefrontal cortex (dmPFC) measured by RNAscope. Red dots show the expression of the neuronal marker RbFox3, and the green dots show the expression of Prdm2 and blue labeling represents DAPI (4′,6-diamidino-2-phenylindole) staining. (d) Prdm2 mRNA expression is decreased after Prdm2 knockdown. (e) Site of virus injection. *P < 0.05; Scrambled versus PRDM2.
Figure 5
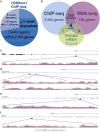
Tripartite approach implicates high confidence targets of PRDM2 in alcohol dependence. (a) Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis identified genomic regions subjected to histone 3 at the lysine 9 residue (H3K9me1) epigenetic regulation in control versus postdependent dorsomedial prefrontal cortex (dmPFC). (b) Overlay of ChIP-seq and RNA-sequencing (RNA-seq) analysis from control versus postdependent dmPFC identified 119 common genes. These analyses were further overlaid with NanoString gene expression analysis from rat mPFC subjected to PRDM2 short hairpin RNA (shRNA) compared with a scramble control (hypergeometric distribution test P = 0.001). (c–f) ChIP-seq data from the four common genes implicated in all three approaches. Top tracks show the UCSC gene relative to the scale shown at the right. The location of the region of differential H3K9me1 enrichment is denoted by the red box below the gene track. The bottom tracks show the Spatial clustering for identification of ChIP-enriched region (SICER)-filtered read enrichment of H3K9me1 ChIP data for the control rats (gray) and postdependent rats (light red) overlaid.
Figure 6
Lentiviral-mediated knockdown of Prdm2 in the dmPFC alters addiction-like behaviors (a–c): Voluntary alcohol-self-administration (a) (One way ANOVA: main effect of group;scrambled versus PRDM2 rats;F(1,40) = 5.9; P < 0.02), compulsive drinking in a quinine adulteration task (b) (Repeated-measure ANOVA: main effect of group;Scrambled versus PRDM2 rats;F(1,48) = 7; P = 0.02), and stress-induced relapse (c) (Repeated-measure ANOVA;main effect of group;scrambled versus PRDM2 rats F(1,84) = 9.3; P = 0.006). Lentiviral knockdown of Prdm2 did not affect control behaviors (d–h) such as locomotor activity (d), average velocity (e), sensitivity to shock intensity (f), sucrose self-administration (g), nor rat weight (h). The mean values (± s.e.m.) of scrambled rats are shown in black bars. The mean values (± s.e.m.) of PRDM2 knockdown rats are shown in gray bars (n = 9–10). *p < 0.05, scrambled versus PRDM2 rats.
Similar articles
- DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity.
Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, Sun H, Schuebel K, Zhou Z, Yuan Q, Vendruscolo LF, Goldman D, Heilig M. Barbier E, et al. J Neurosci. 2015 Apr 15;35(15):6153-64. doi: 10.1523/JNEUROSCI.4571-14.2015. J Neurosci. 2015. PMID: 25878287 Free PMC article. - The GABAB Positive Allosteric Modulator ADX71441 Attenuates Alcohol Self-Administration and Relapse to Alcohol Seeking in Rats.
Augier E, Dulman RS, Damadzic R, Pilling A, Hamilton JP, Heilig M. Augier E, et al. Neuropsychopharmacology. 2017 Aug;42(9):1789-1799. doi: 10.1038/npp.2017.53. Epub 2017 Mar 15. Neuropsychopharmacology. 2017. PMID: 28294133 Free PMC article. - Prefrontal cortex expression of chromatin modifier genes in male WSP and WSR mice changes across ethanol dependence, withdrawal, and abstinence.
Hashimoto JG, Gavin DP, Wiren KM, Crabbe JC, Guizzetti M. Hashimoto JG, et al. Alcohol. 2017 May;60:83-94. doi: 10.1016/j.alcohol.2017.01.010. Epub 2017 Mar 14. Alcohol. 2017. PMID: 28433423 Free PMC article. - [Epigenetic mechanisms and alcohol use disorders: a potential therapeutic target].
Legastelois R, Jeanblanc J, Vilpoux C, Bourguet E, Naassila M. Legastelois R, et al. Biol Aujourdhui. 2017;211(1):83-91. doi: 10.1051/jbio/2017014. Epub 2017 Jul 6. Biol Aujourdhui. 2017. PMID: 28682229 Review. French. - Possible Role of CRF-Hcrt Interaction in the Infralimbic Cortex in the Emergence and Maintenance of Compulsive Alcohol-Seeking Behavior.
Kim JS, Martin-Fardon R. Kim JS, et al. Alcohol Clin Exp Res. 2020 Feb;44(2):354-367. doi: 10.1111/acer.14264. Epub 2020 Jan 2. Alcohol Clin Exp Res. 2020. PMID: 31840823 Free PMC article. Review.
Cited by
- Identification of Schizophrenia Susceptibility Loci in the Urban Taiwanese Population.
Huang CC, Wang YG, Hsu CL, Yeh TC, Chang WC, Singh AB, Yeh CB, Hung YJ, Hung KS, Chang HA. Huang CC, et al. Medicina (Kaunas). 2024 Aug 6;60(8):1271. doi: 10.3390/medicina60081271. Medicina (Kaunas). 2024. PMID: 39202552 Free PMC article. - Brain and Cognition for Addiction Medicine: From Prevention to Recovery Neural Substrates for Treatment of Psychostimulant-Induced Cognitive Deficits.
D'Souza MS. D'Souza MS. Front Psychiatry. 2019 Jul 24;10:509. doi: 10.3389/fpsyt.2019.00509. eCollection 2019. Front Psychiatry. 2019. PMID: 31396113 Free PMC article. Review. - Neurobiological Basis of Aversion-Resistant Ethanol Seeking in C. elegans.
Jee C, Batsaikhan E, Salim C. Jee C, et al. Metabolites. 2022 Dec 31;13(1):62. doi: 10.3390/metabo13010062. Metabolites. 2022. PMID: 36676987 Free PMC article. - Epigenetic drugs and psychedelics as emerging therapies for alcohol use disorder: insights from preclinical studies.
Hilal FF, Jeanblanc J, Deschamps C, Naassila M, Pierrefiche O, Ben Hamida S. Hilal FF, et al. J Neural Transm (Vienna). 2024 May;131(5):525-561. doi: 10.1007/s00702-024-02757-3. Epub 2024 Mar 30. J Neural Transm (Vienna). 2024. PMID: 38554193 Review. - Intermittent Ethanol during Adolescence Leads to Lasting Behavioral Changes in Adulthood and Alters Gene Expression and Histone Methylation in the PFC.
Wolstenholme JT, Mahmood T, Harris GM, Abbas S, Miles MF. Wolstenholme JT, et al. Front Mol Neurosci. 2017 Sep 26;10:307. doi: 10.3389/fnmol.2017.00307. eCollection 2017. Front Mol Neurosci. 2017. PMID: 29018328 Free PMC article.
References
- Meinhardt MW, Sommer WH. Postdependent state in rats as a model for medication development in alcoholism. Addict Biol. 2015;20:1–21. - PubMed
MeSH terms
Substances
Grants and funding
- R01 AA017943/AA/NIAAA NIH HHS/United States
- R01 AA023781/AA/NIAAA NIH HHS/United States
- R21 DA035592/DA/NIDA NIH HHS/United States
- R01 MH084880/MH/NIMH NIH HHS/United States
- P50 NS071674/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases