Fibronectin contributes to notochord intercalation in the invertebrate chordate, Ciona intestinalis - PubMed (original) (raw)
Fibronectin contributes to notochord intercalation in the invertebrate chordate, Ciona intestinalis
Fernando Segade et al. Evodevo. 2016.
Abstract
Background: Genomic analysis has upended chordate phylogeny, placing the tunicates as the sister group to the vertebrates. This taxonomic rearrangement raises questions about the emergence of a tunicate/vertebrate ancestor.
Results: Characterization of developmental genes uniquely shared by tunicates and vertebrates is one promising approach for deciphering developmental shifts underlying acquisition of novel, ancestral traits. The matrix glycoprotein Fibronectin (FN) has long been considered a vertebrate-specific gene, playing a major instructive role in vertebrate embryonic development. However, the recent computational prediction of an orthologous "vertebrate-like" Fn gene in the genome of a tunicate, Ciona savignyi, challenges this viewpoint suggesting that Fn may have arisen in the shared tunicate/vertebrate ancestor. Here we verify the presence of a tunicate Fn ortholog. Transgenic reporter analysis was used to characterize a Ciona Fn enhancer driving expression in the notochord. Targeted knockdown in the notochord lineage indicates that FN is required for proper convergent extension.
Conclusions: These findings suggest that acquisition of Fn was associated with altered notochord morphogenesis in the vertebrate/tunicate ancestor.
Keywords: Chordate evolution; Convergent extension; Extracellular matrix; Fibronectin; Notochord; Tunicates.
Figures
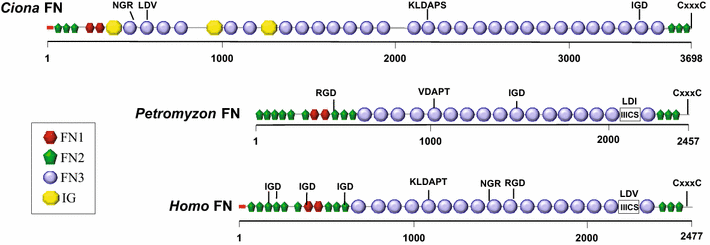
Fig. 1
Comparison of the domain architecture of vertebrate and tunicate FN proteins. Protein domains in full-length Ciona intestinalis FN (3698 amino acids), human FN isoform 1 (accession number NP_001293061; 2477 amino acids) and lamprey (Petromyzon) FN (PMZ_0026015 [19]; fragment, 2457 amino acids) were identified with the SMART 7 domain prediction tool. FN and IG (immunoglobulin) domains as indicated in legend; red box signal peptide sequence; IIICS: type III connecting segment. Potential integrin-binding motifs (including IGD, NGR, KLDAP, DGR, LDV or RGD) and dimerization signal CxxxC are shown above the sequences
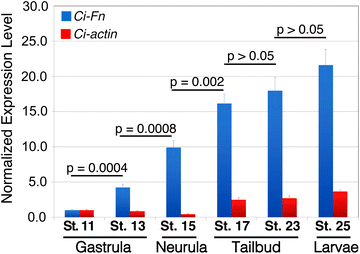
Fig. 2
_Ci_-Fn mRNA accumulation throughout the development of Ciona embryos. Relative expression levels of _Ci_-Fn (blue bars) and _Ci_-actin (red bars) mRNA were obtained by quantitative PCR and normalized by parallel amplification of Ciona 18S target sequences and scaled in relation to minimal expression levels detected at Stage 11. Bars represent the mean ± SD for triplicate samples. P values are for the indicated pairwise comparisons by unpaired t test analysis
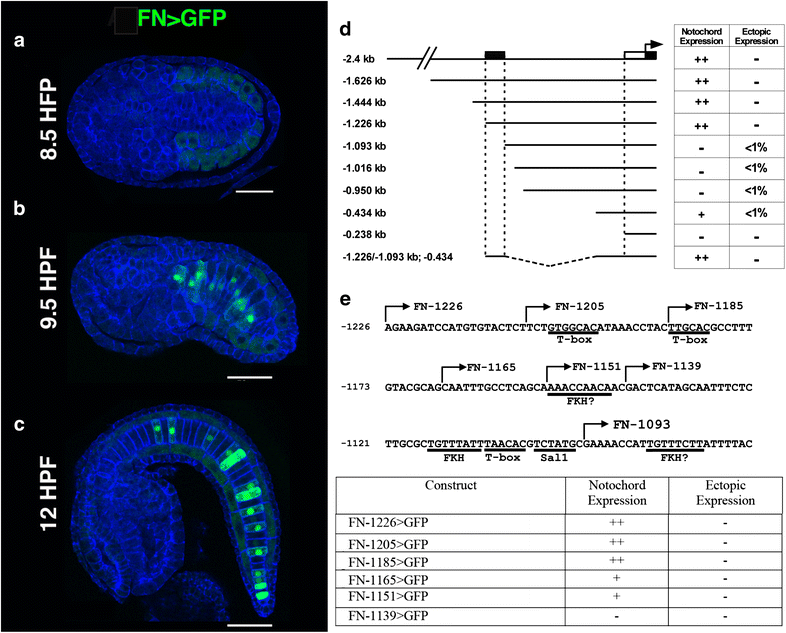
Fig. 3
Reporter analysis of the _Ci_-Fn regulatory element. a–c Representative embryos displaying full-length Fn>GFP reporter expression at three different stages, scale bars = 50 μm. a At 8.5 h post-fertilization (HPF, Stage 17) reporter expression is not detected, background staining (faint green) is observed in developing muscle cells. b Mosaic FN reporter expression in the notochord lineage is initially detected at approximately 9.5 HPF (Stage 18/19). c _Ci_-Fn reporter expression in the notochord continues at later developmental stages as represented by a 12 HFP (Stage 21) embryo. d Schematic illustration of reporter constructs. The 5′ boundary relative to the initiator codon of the _Ci_-Fn sequence in each construct is identified by numbers on the left. Table on the right summarizes observed patterns of GFP expression. Relative abundances were estimated by comparing numerous transgenic embryos (>100 per construct). For ectopic expression, <1 % indicates the detection of GFP expression in non-notochord tissues in less than 1 in 100 embryos examined. Stippled box notochord enhancer region; white rectangle, core promoter; black rectangle 5′ untranslated region. e Map and reporter expression for deletions and mutations within the _Ci_-Fn minimal (−1226, −1093) enhancer region. Putative binding sites are underlined. Arrows indicate the 5′ ends of serial deletion constructs
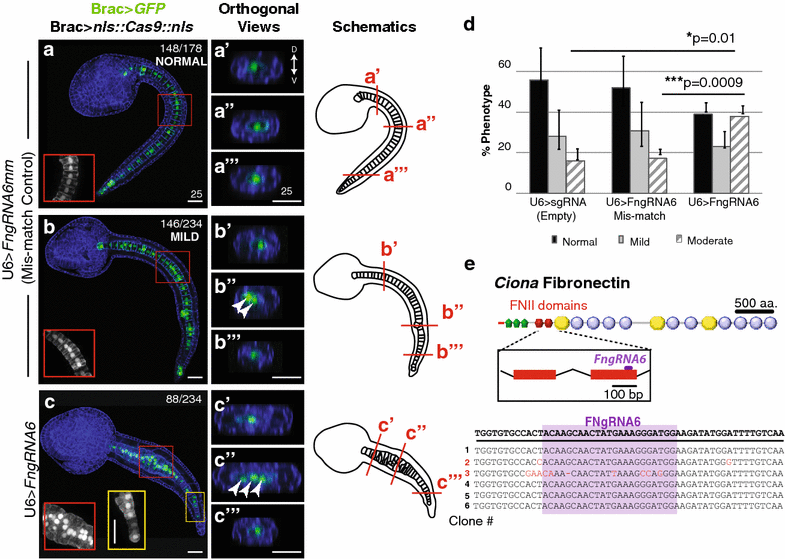
Fig. 4
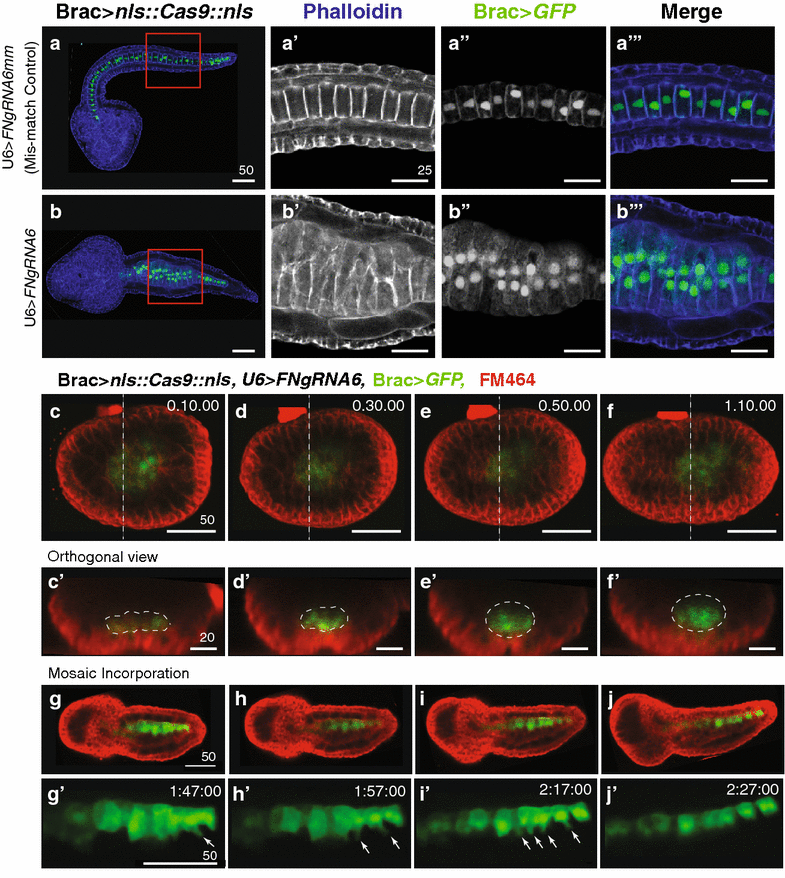
Ciona Fibronectin is necessary for intercalation of notochord cells during convergent extension. a–c′′′ Representative micrographs showing lateral projections and accompanying 2-μm orthogonal sections of notochord cells in late tailbud embryos co-transfected as labeled. Red and yellow boxes indicate location of insets. Approximate locations of orthogonal sections (a′–c′′′) are indicated in the associated schematics. Misaligned cells are indicated in orthogonal sections (arrows). Phalloidin (blue). Note that no reduction in notochord cell numbers occurred in these samples. In all 15 U6>FN6sgRNA embryos with bilateral incorporation of the Bra>GFP, 40 notochord cells were present. d Graphical summary of CRISPR/CAS9 data. Data were obtained from three independent trials, _n_>29/trial. Error bars represent the S.E.M. P values for moderately defective phenotypes in U6>sgRNA (empty) and U6>FN6sgRNA mismatch (controls) versus U6>FN6sgRNA are indicated. P = 0.882 for U6>sgRNA mild versus U6>FNgRNA6 mm mild. Significance was determined using a two-tailed unpaired t test. e Approximate location of gRNA target sequence in FNII domain. Alignment of FN alleles cloned from pooled embryos electroporated with Brac>nls::Cas9::nls and U6>FN6gRNA. Scale bars 25 μm. Embryos oriented anterior to the left
Fig. 5
_Ci_-Fn knockdown specifically blocks the ability of medial protrusions to drive productive notochord intercalation. a, b′′′ Representative micrographs showing sections of phalloidin-stained (blue) late tailbud embryos co-transfected as labeled. Red and boxes (a, b) indicate location of zoomed in images (a′, b′′′). a′, b Representative section displaying typical medial lateral polarity of phalloidin enriched protrusions in non-intercalating cells. a′′, b′′ Representative section displaying typical medial localization of nuclei in non-intercalating cells. c, j′ Still images from movies of embryos co-transfected as labeled. c–f′ Representative embryo with bilateral incorporation of Brac>GFP showing invagination of notochord cells in 4-um orthogonal sections (white outlines, c′–f′) taken from Supplemental Movie 1. Dashed lines in (c–f) indicate location of orthogonal images (c′–f′). g–j′ Representative embryo with unilateral incorporation of Brac>GFP showing medial protrusions of notochord cells (arrows), taken from Supplemental Movie 5. Images represent 10- or 20-min intervals, time stamp in upper right corner. Scale bars in μm. Embryos oriented anterior to the left
Similar articles
- Evolutionary changes in the notochord genetic toolkit: a comparative analysis of notochord genes in the ascidian Ciona and the larvacean Oikopleura.
Kugler JE, Kerner P, Bouquet JM, Jiang D, Di Gregorio A. Kugler JE, et al. BMC Evol Biol. 2011 Jan 20;11:21. doi: 10.1186/1471-2148-11-21. BMC Evol Biol. 2011. PMID: 21251251 Free PMC article. - Genomics reveal ancient forms of stanniocalcin in amphioxus and tunicate.
Roch GJ, Sherwood NM. Roch GJ, et al. Integr Comp Biol. 2010 Jul;50(1):86-97. doi: 10.1093/icb/icq010. Epub 2010 Apr 1. Integr Comp Biol. 2010. PMID: 21558190 - The notochord gene regulatory network in chordate evolution: Conservation and divergence from Ciona to vertebrates.
Di Gregorio A. Di Gregorio A. Curr Top Dev Biol. 2020;139:325-374. doi: 10.1016/bs.ctdb.2020.01.002. Epub 2020 Feb 12. Curr Top Dev Biol. 2020. PMID: 32450965 Review. - CRISPR/Cas9 Protocols for Disrupting Gene Function in the Non-vertebrate Chordate Ciona.
Popsuj S, Cohen L, Ward S, Lewis A, Yoshida S, Herrera RA, Cota CD, Stolfi A. Popsuj S, et al. Integr Comp Biol. 2024 Nov 21;64(5):1182-1193. doi: 10.1093/icb/icae108. Integr Comp Biol. 2024. PMID: 38982335 Review. - Key steps in the morphogenesis of a cranial placode in an invertebrate chordate, the tunicate Ciona savignyi.
Kourakis MJ, Newman-Smith E, Smith WC. Kourakis MJ, et al. Dev Biol. 2010 Apr 1;340(1):134-44. doi: 10.1016/j.ydbio.2010.01.016. Epub 2010 Jan 22. Dev Biol. 2010. PMID: 20096682 Free PMC article.
Cited by
- Architectural delineation and molecular identification of extracellular matrix in ascidian embryos and larvae.
Wei J, Wang G, Li X, Ren P, Yu H, Dong B. Wei J, et al. Biol Open. 2017 Sep 15;6(9):1383-1390. doi: 10.1242/bio.026336. Biol Open. 2017. PMID: 28916708 Free PMC article. - Cis-regulatory interfaces reveal the molecular mechanisms underlying the notochord gene regulatory network of Ciona.
Negrón-Piñeiro LJ, Wu Y, Popsuj S, José-Edwards DS, Stolfi A, Di Gregorio A. Negrón-Piñeiro LJ, et al. Nat Commun. 2024 Apr 8;15(1):3025. doi: 10.1038/s41467-024-46850-3. Nat Commun. 2024. PMID: 38589372 Free PMC article. - CRISPR Knockouts in Ciona Embryos.
Gandhi S, Razy-Krajka F, Christiaen L, Stolfi A. Gandhi S, et al. Adv Exp Med Biol. 2018;1029:141-152. doi: 10.1007/978-981-10-7545-2_13. Adv Exp Med Biol. 2018. PMID: 29542087 Free PMC article. Review. - The ion channel Anoctamin 10/TMEM16K coordinates organ morphogenesis across scales in the urochordate notochord.
Liang Z, Dondorp DC, Chatzigeorgiou M. Liang Z, et al. PLoS Biol. 2024 Aug 22;22(8):e3002762. doi: 10.1371/journal.pbio.3002762. eCollection 2024 Aug. PLoS Biol. 2024. PMID: 39173068 Free PMC article. - The Ascidia Ciona robusta Provides Novel Insights on the Evolution of the AP-1 Transcriptional Complex.
Marotta P, Salatiello F, Ambrosino L, Berruto F, Chiusano ML, Locascio A. Marotta P, et al. Front Cell Dev Biol. 2021 Aug 3;9:709696. doi: 10.3389/fcell.2021.709696. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34414189 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous