On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models - PubMed (original) (raw)
. 2016 Nov 3;539(7627):107-111.
doi: 10.1038/nature19795. Epub 2016 Sep 5.
Xinlin Du 2, James P Rizzi 2, Ella Liberzon 1, Abhishek A Chakraborty 1, Wenhua Gao 1, Ingrid Carvo 1 3, Sabina Signoretti 1 3, Richard K Bruick 4, John A Josey 2, Eli M Wallace 2, William G Kaelin 1 5
Affiliations
- PMID: 27595393
- PMCID: PMC5499381
- DOI: 10.1038/nature19795
On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models
Hyejin Cho et al. Nature. 2016.
Abstract
Clear cell renal cell carcinoma, the most common form of kidney cancer, is usually linked to inactivation of the pVHL tumour suppressor protein and consequent accumulation of the HIF-2α transcription factor (also known as EPAS1). Here we show that a small molecule (PT2399) that directly inhibits HIF-2α causes tumour regression in preclinical mouse models of primary and metastatic pVHL-defective clear cell renal cell carcinoma in an on-target fashion. pVHL-defective clear cell renal cell carcinoma cell lines display unexpectedly variable sensitivity to PT2399, however, suggesting the need for predictive biomarkers to be developed to use this approach optimally in the clinic.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
X.D., J.P.R., R.B., J.A.J, E.M.W., and W.G.K. own equity in Peloton as Peloton employees (X.D., J.P.R., J.A.J., and E.M.W), licensors (R.B), or advisors (R.B. and W.G.K.).
Figures
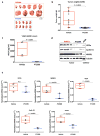
Extended Data Figure 1. Binding of PT2399 to PAS-B domain of human HIF2α as determined by X-Ray co-crystal structure
a, X-Ray co-crystal of PT2399 (magenta) bound to HIF2α/ARNT PAS-B domains (ARNT removed for clarity). b, X-Ray co-crystal of PT2399 (magenta) with HIF2α/ARNT PAS-B domains (zoomed in on HIF2α PAS-B pocket). c, Immunoblots of anti-ARNT1 immunoprecipitates (IP) of Hep3B cells treated with PT2399 or DMSO. d, Immunoblot of 786-O cells expressing shRNA against HIF2α (3806) or control shRNA. e, HIF2α specific gene regulation in Hep3B; n=3 biological replicates. f, Immunoblot analysis (top) and quantification (bottom) of HIF2α in 786-O cells treated with DMSO or PT2399 for 16 hours and then exposed to cycloheximide for the indicated time periods; n=3 biological replicates. g, Enrichment plots for representative gene sets previously linked to HIF, hypoxia, or c-Myc. h, i, Plasma PT2399 levels after administration of a single dose of PT2399 to CD-1 mice; n=3 per time point from one experiment.
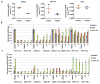
Extended Data Figure 2. Inhibition of cell proliferation by PT2399 ex vivo
a, b, Immunoblot analysis of 786-O cells after CRISPR-based gene editing with control sgRNA or HIF2α sgRNA (guides 4 and 6). In (b) cells were also infected with an empty vector (EV) or a virus expressing an HIF2α sgRNA guide 6-resistant HIF2α cDNA. c–f, Cell proliferation of parental 786-O cells (c) and 786-O clones subjected to CRISPR-based gene editing with a control sgRNA (d) or in which endogenous HIF2α was successfully inactivated using two different HIF2α sgRNAs (guides 4 and 6) (e, f); n=3 biological replicates. g–j, Proliferation curves for 786- M1A cells (g), UMRC-2 (h), Caki-1 cells (i), and Caki-2 cells (j) treated with the indicated concentrations of PT2399. k, Immunoblot analysis of the indicated cell lines. l, m, Proliferation curves for MDA-MB-231 (l) and A549 cells (m) treated with indicated concentrations of PT2399; n=3 biological replicates. Data as mean ± s.d. (c–j, l and m)
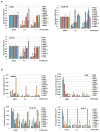
Extended Data Figure 3. Effects of PT2399 on soft agar growth
a–c, Soft agar colonies formed by 786-O cells (a), A498 cells (b), and the indicated cell lines (c) in the presence of PT2399 at the indicated concentrations for 21 days; n=3 biological replicates. d, Soft agar colonies formed by the indicated polyclonal cell line populations after CRISPR-based gene editing with control sgRNA or HIF2α sgRNA (guides 4 and 6); n=3 biological replicates. The reason for the differential sensitivity of RCC10 cells to PT2399 and the HIF2α sgRNAs is not yet clear. e, Immunoblot analysis of the cells used in (d). For SLR21 cells, 1mM DMOG was treated for 16 hrs to detect HIF2α. f, g, Quantification of soft agar colonies formed in (a–d and Fig. 5h, j), respectively; n=3. Data as mean ± s.e.m. *P<0.01 by two-tailed Student’s _t_-tests (f, g).
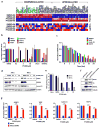
Extended Data Figure 4. Pharmacodynamic effects of PT2399 in vivo
a, Levels of the indicated mRNAs, normalized to beta actin, in 786-O orthotopic tumors treated with PT2399 30 mg/kg (n =3 mice from two independent experiments) or vehicle (n=3 mice from two independent experiments) twice daily for two days in vivo. b, Immunoblot analysis of 786-O orthotopic tumors treated with PT2399 30 mg/kg or vehicle twice daily for two days in vivo; for vehicle, n=3 mice from two independent experiments and PT2399, n=3 mice from two independent experiments. c, Quantification of Ki-67 staining (vehicle, n=3; PT2399, n=3 mice from two independent experiments) d, Immunohistochemistry of representative 786-O orthotopic tumors treated with PT2399 30 mg/kg or vehicle twice daily for two days in vivo; for vehicle, n=3 and PT2399, n=3. Scale bars, 50 μm. e, Microvessel Density (vehicle, n=5; PT2399, n=3 mice from two independent experiments) from tumors as in (d). f, g, Representative tumors at necropsy (f) and serum VEGF concentrations (vehicle, n=10; PT2399, n=11 mice from three independent experiments) (g) from mice as in Fig. 4a–c just prior to necropsy. h, Representative immunohistochemical staining of 786-O tumors treated as in Fig. 4a–c (vehicle, n=4; PT2399, n=5); Scale bars, 50 μm. Data as median with range (a, c, e and g). Statistical significance was assessed by using Mann-Whitney test (e) or Unpaired _t_-test (g). N.S., _P_>0.05.
Extended Data Figure 5. Antitumor activity of PT2399 in lung colonization and PDX models
a, BLI of lung colonies formed after tail vein injection of 9,000 786-M2A cells treated with PT2399 30 mg/kg or vehicle twice daily by oral gavage. Treatment began at week 1. b, Quantification of BLI values as in (a). Data as median with range (Vehicle, n=2 and PT2399, n=3 mice from one experiment). c, Partial rescue of PT2399 pharmacodynamic effect by HIF2α S304M; n=3 biological replicates Levels of the indicated mRNAs, normalized to beta actin mRNA and then to DMSO treatment, in cells from Fig. 4g treated with PT2399 at the indicated concentrations for 48 hours; n=3. Data as mean ± s.e.m. *P<0.05 by two-tailed Student’s _t_-tests. Note that rescue is only partial, perhaps because these cells still produce endogenous wild-type HIF2α in addition to exogenous HIF2α S304M. d, Subcutaneous PDX measurements in mice randomized to the indicated treatments, including the FDA-approved ccRCC drug sunitinib, when the tumors reached 200 to 300 mm3. P<0.05 for difference between PT2399 and vehicle (n = 8, unpaired t-test). e, Immunohistochemistry of PDX in (d) before treatment. Scale bars, 100 μm. Data as mean ± s.e. m. (d).
Extended Data Figure 6. Antitumor activity of PT2399 using A498 cells
a, b, Representative BLI (a) and quantification of BLI measurements (b) of orthotopic tumors formed by A498 cells expressing firefly luciferase under the control of CMV promoter before and after (30 days) treatment with PT2399 30 mg/kg or vehicle twice daily by oral gavage (vehicle, n=10 and PT2399, n=10 mice from two independent experiments) c, d, Representative tumors (c) and tumor masses (d) at necropsy from mice treated as in (a) (vehicle, n=10 and PT2399, n=10 mice). e, Serum VEGF concentrations from mice treated as in (a) at time of necropsy (vehicle, n=4; PT2399, n=4 mice from two independent experiments). f, Immunoblot of representative tumors from (a); for vehicle and PT2399, n=4 and n=3, respectively. g, Levels of the indicated mRNAs, normalized to beta actin, in A498 orthotopic tumors (vehicle, n=4; PT2399, n=3 mice from two independent experiments) treated as in (a). Data as median with range. Statistical significance was assessed by using two-tailed Student’s _t_-tests with Welch’s correction (b and d) or Mann-Whitney test (d and e). N.S., _P_>0.05.
Extended Data Figure 7. Elimination of HIF1α does not render UMRC-2 cells sensitive to PT2399 in soft agar assays
a–c, Firefly luciferase activity in the indicated cell lines after infection with a virus containing firefly luciferase under the control of a HIF-responsive (HRE-Luc) promoter (a, c) or CMV promoter (b) and treatment with the indicated concentrations of PT2399 for 16 hrs relative to DMSO-treated controls; n=3 biological replicates. d, Immunoblot analysis of HRE-Luc expressing UMRC-2 cells after CRISPR-based gene editing with control sgRNA or HIF1α sgRNA (guides 2 and 3). Note that deletion of HIF1α in (c) and (d) was used to eliminate the contribution of HIF1α in (a). e, f, Immunoblot (e) and mRNA levels (f) of UMRC-2 cells after CRISPR-based gene editing with control sgRNA or HIF1α sgRNA (guides 2 and 3). In (f) mRNA levels were normalized to beta-actin and then to the corresponding control sgRNA value; n=3 biological replicates. g, Soft agar assays of the cells analyzed in (e) and (f) in the presence of the indicated concentrations of PT2399; n=3 biological replicates. Data as mean ± s.e.m. (a–c and f).
Extended Data Figure 8. Variable sensitivity of ccRCC lines to PT2399 and pVHL
a, Levels of the indicated mRNAs, normalized to beta actin, in UMRC-2 orthotopic tumors treated with PT2399 30 mg/kg or vehicle twice daily for 1 month; for vehicle, n=2; PT2399, n=2 mice from one experiment. b, c, Downregulation of HIF-responsive mRNAs by PT2399 in indicated cell lines. For each cell line the mRNA levels were normalized to beta actin mRNA (c) and then normalized to untreated value for that cell line (b); n=3 biological replicates. Data as median with range (a) and mean ± s.d. (b, c). d–e, Variable suppression of HIF target genes by PT2399 across a panel of ccRCC cell lines. Downregulation of HIF-responsive VEGF, Cyclin D1, and GLUT1 mRNAs by PT2399 in the indicated cell lines. For each cell line the mRNA levels were normalized to beta actin (e) and then normalized to untreated value for that cell line (d); n=3 biological replicates. SLR21 VHL+/+ renal carcinoma cell was included for comparison. Data as mean ± s.d.
Extended Data Figure 9. HIF2 dependence of RCC10 cells
a, Immunoblot analysis of anti-ARNT1 immunoprecipitates (IP) and whole cell extracts (input) prepared from RCC10 cells treated with increasing amounts of PT2399 or DMSO. Control IP without ARNT1 antibody was marked as ‘C’. b, Levels of the indicated mRNAs, normalized to beta actin, in 786-O, A498 and RCC10 cells treated with PT2399 at the indicated concentration for 24 hrs or an effective HIF2α sgRNA (sgHIF2α-6), and then normalized to cells treated with DMSO or a control sgRNA, respectively. Data as mean ± s.d.; n=3 biological replicates. c, immunoblot analysis of RCC10 cells after CRISPR based editing with HIF2α sgRNAs or control sgRNA. d, e, soft agar colonies formed by RCC10 cells as in (c); n=3 biological replicates. In (e) cells were engineered to express an exogenous sgRNA-resistant HIF2α or empty vector (EV);n=3. f, Soft agar colony counts as in (e) using ImageJ software. Colonies were counted using the following criteria: circularity range from 0.5 to 1.0 and size (pixels2) from 200 to infinity. Data shown mean ± s.e.m. Statistical significance was assessed by using two-tailed Student’s t_-tests (f). *P<0.05. g–k, p53 pathway status in ccRCC lines. g, j, Immunoblot analysis of the indicated cell lines treated for 16 hours with etoposide or vehicle. Note overproduction of p53 in RCC10 cells and off-size p53 band in UMRC-2 cells. SLR21 cells are VHL+/+. Red = PT2399 sensitive in soft agar assays. Blue = PT2399 insensitive. RCC4 cells do not form soft agar colonies and are therefore indeterminate. h, Immunoblot analysis of 786-O cells that were infected with an empty lentivirus conferring puromycin resistance and then later found to have spontaneously acquired a p53 mutation (R248W) compared to cells that retained wild-type p53. Cells were treated with PT2399 for 48 hours or with nutlin-3 (30 μM) or etoposide (20 μM) for 16 hrs. i, soft agar colony formation from cells in (h) treated with PT2399; n=3 biological replicates. k, Immunoblot analysis of parental 786-O cells that underwent CRISPR-based gene editing with a control sgRNA or HIF2α sgRNA (guide 6) (as in Extended Data Fig. 2_a) then treated with PT2399 for 58 hours or treated with nutlin-3 (30 μM) or etoposide (20 μM) for 10 hrs.
Extended Data Figure 10. Loss of HIF2α does not suppress UMRC-2 orthotopic tumor growth
a–c, Tumors (a), tumor weights (b), and tumor immunoblots (c) at necropsy from mice after orthotopic injection of UMRC-2 cells that had undergone CRISPR-based editing with sgControl or sgHIF2α-6 as in Fig. 5g; sgCon, n=5 and sgHIF2α, n=5 mice from two independent experiments. The reason for the variable HIF2α levels in (c) is unknown but could reflect, at least partly, variable amounts of host-derived cells in the tumor samples. d, Levels of the indicated mRNAs, normalized to beta actin, in tumors from a–c. Data as median with range (b, d). Statistical significance was assessed by using two-tailed Student’s _t_-tests (b) or Mann-Whitney test (d). Loss of HIF2α did suppress subcutaneous tumor growth (data not shown).
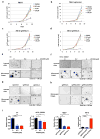
Figure 1
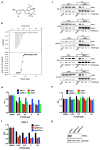
PT2399 downregulates HIF target genes. a, Binding of PT2399 (inset) to human HIF2α PAS-B domain assayed by isothermal calorimetry (n=2) b, Immunoblots of anti-ARNT1 immunoprecipitates (IP) and whole cell extracts (input) from ccRCC cell lines treated with PT2399 or DMSO. C = control IP without ARNT1 antibody. c, Heat map of changes in mRNAs in 786-O cells [parental and HIF2α−/− generated with sgHIF2α (guide #6)] treated with 2 μM PT2399 or DMSO for indicated duration (hours). n=1. Examples of HIF target genes are indicated in green. d, Venn diagram of gene sets regulated by PT2399. 249 gene sets were regulated in parental 786-O cells, but not in 786-O HIF2α −/− cells. e, f, Levels of the indicated mRNAs, normalized to beta actin, and then to DMSO controls, in 786-O cells treated with PT2399 at the indicated concentration for 24 hrs (e) or duration with 2 μM PT2399 (f). 786-O cells expressing a HIF2α shRNA or control shRNA were included in (e) for comparison; n=3. g, Immunoblots of 786-O cells treated with PT2399. h, VEGF concentration, normalized to total cellular protein, in media conditioned by 786-O cells treated with PT2399; n=3 biological replicates. Data as mean ± s.e.m. (e and f) or mean ± s.d. (h)
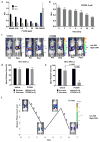
Figure 2
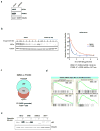
On-target effects of PT2399 on transcription and soft agar growth. a, Immunoblots of HIF2α_−_/− 786-O cells (sgHIF2α #6) infected with a lentivirus encoding V5-tagged HIF2α (wild-type), HIF2α (S304M), or empty vector. b, Levels of the indicated mRNAs, normalized to beta actin mRNA, and then to DMSO treatment, in cells from (a) treated with PT2399 or DMSO for 48 hours; n=3. c, d, Soft agar colonies formed by 786-O cells infected with a lentivirus encoding HIF2α S304M (d) or the empty vector (c) in the presence of PT2399 at the indicated concentrations for 21 days; n=3 biological replicates. e, f, Soft agar colonies formed by 786-O clones as in (Extended Data Figure 2a, b); n=3 biological replicates. In f, the 786-O HIF2α−/− cells (sgHIF2α #6) were superinfected with a lentivirus encoding a sgRNA-resistant HIF2α cDNA (HIF2α) or with the empty vector (EV). g, Quantification of soft agar colony formation in (c–f). Data as mean ± s.e.m. *P<0.05, **P<0.01 and ***P<0.001 by two-tailed Student’s _t_-tests (b, g).
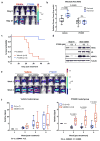
Figure 3
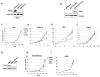
PT2399 pharmacodynamic effects in vivo. a, b Light emission, normalized to total cellular protein (a) and cell number (b) in 786-O 3XHRE-Luc reporter cells expressing VHL or an empty vector (Vec). The cells were treated with PT2399 overnight (a) or at 2 μM for the indicated durations (b). 1 mM DMOG was included in (a) as a control; n=3. c, Luc values of 786-O derivatives expressing Luc driven by the indicated promoters and then exposed to either PT2399 or to 1 mM DMOG; n=3 biological replicates. d, Representative bioluminescent images (BLI) of mice with orthotopic tumors formed by 786-O 3XHRE-Luc reporter cells (left kidney) or 786-O CMV-Luc reporter cells (right kidney). Images were obtained before and after two days of PT2399 30 mg/kg given twice daily (n=6 mice from two independent experiments) or vehicle (n=6 mice from two independent experiments) by oral gavage. e, f, Quantification of BLI from mice as in (d); n=4 mice from two independent experiments. Values were normalized to the pretreatment values for each treatment. g, Serial BLI of a mouse as in (d) treated with PT2399 (blue arrows). Shown on the y-axis are absolute photon counts for the 3XHRE-Luc tumor. Data as mean ± s.e.m. (a–c, e and f). Statistical significance was assessed using Mann-Whitney tests (f).
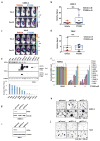
Figure 4
PT2399 antitumor activity. a, Representative BLI of orthotopic tumors formed by 786-O CMV-Luc cells before and after PT2399 30 mg/kg or vehicle twice daily by oral gavage for 30 days (vehicle, n=15 mice from three independent experiments and PT2399, n=13 mice from three independent experiments) b, Spider plot showing growth of tumors as in (a) as determined by serial BLI. For each tumor the BLI values were normalized to the corresponding day 0 value. c, Quantification of BLI from mice as in (a). For c, the value for each tumor was normalized to the pretreatment value for that tumor. d, Representative BLI of lung colonies formed by M2A-Luc cells and treated with PT2399 30 mg/kg or vehicle twice daily by oral gavage (vehicle, n=9; PT2399, n=9 from two independent experiments). e, f, Quantification of BLI values (e) and Kaplan-Meier survival curves (f) from mice treated as in (d) (vehicle, n=9 mice from two independent experiments and PT2399, n=9 mice from two independent experiments). g, Immunoblots of M2A cells infected with a lentivirus encoding HIF2α S304M or empty vector. h, Representative BLI of lung colonies formed by M2A cells in (g) treated with PT2399 30 mg/kg or vehicle twice daily by oral gavage (for EV, vehicle and PT2399, n=7 and n=8, respectively; for S304M, vehicle and PT2399, n=9 and 10, respectively). i, Quantification of BLI values as in (h); for EV, vehicle and PT2399, n=7 and n=8, respectively; for S304M, vehicle and PT2399, n=9 and 10, respectively from two independent experiments. Data as median with range(c, e and i) Statistical significance was assessed using Mann-Whitney test (c and i ), unpaired t-test (e) or log-rank test (f).
Figure 5
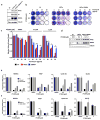
Variable sensitivity of ccRCC lines to PT2399. a, c, Representative BLI of orthotopic tumors formed by UMRC-2 CMV-Luc cells (a) (vehicle, n=8 and PT2399, n=9 mice from two independent experiments) and 769-P CMV-Luc cells (c) (vehicle, n=7 and PT2399, n=8 mice from two independent experiments) before and after (30 and 35 days, respectively) treatment with PT2399 30 mg/kg or vehicle by oral gavage. b, d, Quantification of BLI as in (a) and (c), respectively. e, Immunoblots ccRCC cells lines, including quantification of HIF2α protein levels, normalized to tubulin. HIF1α in A498 and SKRC20 cells is mutated and defective. f, NDRG1 mRNA levels in ccRCC cell lines treated with PT2399 for 24 hrs; n=3 biological replicates. For each cell line, after normalization to beta actin, NDRG1 mRNA levels were normalized to untreated value for that cell line. SLR21 VHL+/+ renal carcinoma cells were included in (e) and (f) for comparison. g, i, Immunoblots of UMRC-2 cells (g) and 769-P cells (i) after CRISPR-based gene editing with control sgRNA or HIF2α sgRNA (guides 4 and 6). h, j, Soft agar colonies formed by cells as in (g) and (i); n=3 biological replicates. Data as median with range (b, d) and mean ± s.d. (f). Statistical analysis was performed by using two-tailed Student’s _t_-tests (b, d). N.S., _P_>0.05.
Figure 6
Comment in
- Kidney cancer: HIF-2α - a new target in RCC.
Thoma C. Thoma C. Nat Rev Urol. 2016 Nov;13(11):627. doi: 10.1038/nrurol.2016.184. Epub 2016 Sep 27. Nat Rev Urol. 2016. PMID: 27670614 No abstract available. - Targeting HIF2α in Clear-Cell Renal Cell Carcinoma.
Ricketts CJ, Crooks DR, Linehan WM. Ricketts CJ, et al. Cancer Cell. 2016 Oct 10;30(4):515-517. doi: 10.1016/j.ccell.2016.09.016. Cancer Cell. 2016. PMID: 27728802 - Basic/Translational Science Survey Section: Kidney Cancer.
Maranchie JK. Maranchie JK. Urol Oncol. 2017 Apr;35(4):157. doi: 10.1016/j.urolonc.2017.01.008. Epub 2017 Feb 15. Urol Oncol. 2017. PMID: 28214282 No abstract available.
Similar articles
- Targeting renal cell carcinoma with a HIF-2 antagonist.
Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, Homayoun F, Ma Y, Patel N, Yell P, Hao G, Yousuf Q, Joyce A, Pedrosa I, Geiger H, Zhang H, Chang J, Gardner KH, Bruick RK, Reeves C, Hwang TH, Courtney K, Frenkel E, Sun X, Zojwalla N, Wong T, Rizzi JP, Wallace EM, Josey JA, Xie Y, Xie XJ, Kapur P, McKay RM, Brugarolas J. Chen W, et al. Nature. 2016 Nov 3;539(7627):112-117. doi: 10.1038/nature19796. Epub 2016 Sep 5. Nature. 2016. PMID: 27595394 Free PMC article. - Targeting HIF-2 α in clear cell renal cell carcinoma: A promising therapeutic strategy.
Martínez-Sáez O, Gajate Borau P, Alonso-Gordoa T, Molina-Cerrillo J, Grande E. Martínez-Sáez O, et al. Crit Rev Oncol Hematol. 2017 Mar;111:117-123. doi: 10.1016/j.critrevonc.2017.01.013. Epub 2017 Jan 28. Crit Rev Oncol Hematol. 2017. PMID: 28259286 Review. - HIF-independent synthetic lethality between CDK4/6 inhibition and VHL loss across species.
Nicholson HE, Tariq Z, Housden BE, Jennings RB, Stransky LA, Perrimon N, Signoretti S, Kaelin WG Jr. Nicholson HE, et al. Sci Signal. 2019 Oct 1;12(601):eaay0482. doi: 10.1126/scisignal.aay0482. Sci Signal. 2019. PMID: 31575731 Free PMC article. - Sensitivity of VHL mutant kidney cancers to HIF2 inhibitors does not require an intact p53 pathway.
Stransky LA, Vigeant SM, Huang B, West D, Denize T, Walton E, Signoretti S, Kaelin WG Jr. Stransky LA, et al. Proc Natl Acad Sci U S A. 2022 Apr 5;119(14):e2120403119. doi: 10.1073/pnas.2120403119. Epub 2022 Mar 31. Proc Natl Acad Sci U S A. 2022. PMID: 35357972 Free PMC article. - Allosteric inhibition of HIF-2α as a novel therapy for clear cell renal cell carcinoma.
Yu Y, Yu Q, Zhang X. Yu Y, et al. Drug Discov Today. 2019 Dec;24(12):2332-2340. doi: 10.1016/j.drudis.2019.09.008. Epub 2019 Sep 18. Drug Discov Today. 2019. PMID: 31541711 Review.
Cited by
- Metadynamics-Based Approaches for Modeling the Hypoxia-Inducible Factor 2α Ligand Binding Process.
Callea L, Bonati L, Motta S. Callea L, et al. J Chem Theory Comput. 2021 Jul 13;17(7):3841-3851. doi: 10.1021/acs.jctc.1c00114. Epub 2021 Jun 3. J Chem Theory Comput. 2021. PMID: 34082524 Free PMC article. - Toward a CRISPR-based mouse model of _Vhl_-deficient clear cell kidney cancer: Initial experience and lessons learned.
Stransky LA, Gao W, Schmidt LS, Bi K, Ricketts CJ, Ramesh V, James A, Difilippantonio S, Ileva L, Kalen JD, Karim B, Jeon A, Morgan T, Warner AC, Turan S, Unite J, Tran B, Choudhari S, Zhao Y, Linn DE, Yun C, Dhandapani S, Parab V, Pinheiro EM, Morris N, He L, Vigeant SM, Pignon JC, Sticco-Ivins M, Signoretti S, Van Allen EM, Linehan WM, Kaelin WG Jr. Stransky LA, et al. Proc Natl Acad Sci U S A. 2024 Oct 8;121(41):e2408549121. doi: 10.1073/pnas.2408549121. Epub 2024 Oct 4. Proc Natl Acad Sci U S A. 2024. PMID: 39365820 Free PMC article. - The von Hippel-Lindau Tumor Suppressor Gene: Implications and Therapeutic Opportunities.
Elias R, Zhang Q, Brugarolas J. Elias R, et al. Cancer J. 2020 Sep/Oct;26(5):390-398. doi: 10.1097/PPO.0000000000000480. Cancer J. 2020. PMID: 32947307 Free PMC article. Review. - MK-6482 as a potential treatment for von Hippel-Lindau disease-associated clear cell renal cell carcinoma.
Hasanov E, Jonasch E. Hasanov E, et al. Expert Opin Investig Drugs. 2021 May;30(5):495-504. doi: 10.1080/13543784.2021.1925248. Epub 2021 May 20. Expert Opin Investig Drugs. 2021. PMID: 33945366 Free PMC article. Review. - VHL synthetic lethality screens uncover CBF-β as a negative regulator of STING.
Bertlin JAC, Pauzaite T, Liang Q, Wit N, Williamson JC, Sia JJ, Matheson NJ, Ortmann BM, Mitchell TJ, Speak AO, Zhang Q, Nathan JA. Bertlin JAC, et al. bioRxiv [Preprint]. 2024 Sep 6:2024.09.03.610968. doi: 10.1101/2024.09.03.610968. bioRxiv. 2024. PMID: 39282259 Free PMC article. Preprint.
References
- Kaelin W. Kidney Cancer: Principles and Practice. Vol. 3. Springer International Publishing; 2015. pp. 31–57.
- DIXON DD, et al. ARYL ETHERS AND USES THEREOF. 2015035223. WO. 2015
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical