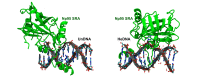Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1 - PubMed (original) (raw)
doi: 10.7554/eLife.17101.
Evan M Cornett 3, Dennis Goldfarb 4, Paul A DaRosa 5, Zimeng M Li 6, Feng Yan 7, Bradley M Dickson 3, Angela H Guo 1, Daniel V Cantu 1, Lilia Kaustov 8, Peter J Brown 8, Cheryl H Arrowsmith 8, Dorothy A Erie 9, Michael B Major 4 7, Rachel E Klevit 5, Krzysztof Krajewski 1, Brian Kuhlman 1 2, Brian D Strahl 1 2, Scott B Rothbart 3
Affiliations
- PMID: 27595565
- PMCID: PMC5012860
- DOI: 10.7554/eLife.17101
Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1
Joseph S Harrison et al. Elife. 2016.
Abstract
The epigenetic inheritance of DNA methylation requires UHRF1, a histone- and DNA-binding RING E3 ubiquitin ligase that recruits DNMT1 to sites of newly replicated DNA through ubiquitylation of histone H3. UHRF1 binds DNA with selectivity towards hemi-methylated CpGs (HeDNA); however, the contribution of HeDNA sensing to UHRF1 function remains elusive. Here, we reveal that the interaction of UHRF1 with HeDNA is required for DNA methylation but is dispensable for chromatin interaction, which is governed by reciprocal positive cooperativity between the UHRF1 histone- and DNA-binding domains. HeDNA recognition activates UHRF1 ubiquitylation towards multiple lysines on the H3 tail adjacent to the UHRF1 histone-binding site. Collectively, our studies are the first demonstrations of a DNA-protein interaction and an epigenetic modification directly regulating E3 ubiquitin ligase activity. They also define an orchestrated epigenetic control mechanism involving modifications both to histones and DNA that facilitate UHRF1 chromatin targeting, H3 ubiquitylation, and DNA methylation inheritance.
Keywords: DNA methylation; UHRF1; biochemistry; biophysics; epigenetics; histone post-translational modifications; human; structural biology; ubiquitin; ubiquitylation.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
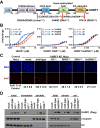
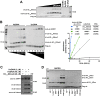
Figure 1.. UHRF1 binding to HeDNA is required for DNA methylation regulation but is dispensable for chromatin interaction.
(A) Domain map of human UHRF1 with identified biochemical functions (top) and loss-of-function point mutations used in this study (bottom; see also Figure 1—figure supplement 1). UBL (ubiquitin-like); TTD (tandem Tudor domain); PHD (plant homeodomain); SRA (SET and RING-associated domain); RING (really interesting new gene). Amino acid positions demarcating domain boundaries are also shown. (B) FP binding assays quantifying the interaction of wild-type, DNAmut, and HeDNAmut MBP-tagged UHRF1 with the indicated FAM-labeled DNA oligonucleotides. Error is represented as ± s.e.m. for two independent experiments. (C) Representative immunofluorescence staining for 5-methylcytosine (5mC) in control and UHRF1 knockdown Hela cells after genetic complementation with the indicated wild-type and mutant forms of full-length UHRF1. Error is represented as ± S.D. from at least four fields of view. Mock, no DNA control; Scale bar, 20 μm. (D) Chromatin association assays for FLAG-tagged UHRF1 (wild-type) or the indicated mutants from asynchronously growing HeLa cells. Mock, no DNA control. DOI:
http://dx.doi.org/10.7554/eLife.17101.003
Figure 1—figure supplement 1.. UHRF1 mutations characterized in this study.
(A) Table of mutant UHRF1 proteins characterized in this study, their functional consequences, and references associated with their initial characterization. (B) Crystal structures of the UHRF1 TTD-PHD domain bound to an H3K9me3 peptide (PDB:3ASK), the UHRF1 SRA domain bound to an HeDNA oligonucleotide (PDB:3CLZ), and the RING domain (PDB:3FLZ). Insets highlight structural details of these interactions that involve residues mutated in this study. DOI:
http://dx.doi.org/10.7554/eLife.17101.004
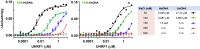
Figure 1—figure supplement 2.. The DNA binding affinity of UHRF1 is highly sensitive to salt concentration.
FP binding assays quantifying the interactions of MBP-UHRF1 with FAM-labeled HeDNA or UnDNA oligonucleotide probes. Error is represented as ± s.e.m. for two independent experiments. DOI:
http://dx.doi.org/10.7554/eLife.17101.005
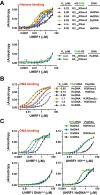
Figure 2.. The DNA- and histone-binding domains of UHRF1 are regulated by reciprocal positive allostery.
(A) FP binding assays quantifying the interaction of MBP-UHRF1 with a C-terminally FAM-labeled H31-20K9me3 peptide (see Supplementary file 1 for a full list of peptides used in this study) in the absence or presence of the indicated unlabeled DNA oligonucleotides. Error is represented as ± s.e.m. for two independent experiments. (B–C) FP binding assays quantifying the interactions of wild-type and the indicated mutant MBP-UHRF1 proteins with FAM-labeled HeDNA or UnDNA in the presence and absence of the indicated unlabeled H31-20 peptides. Error is represented as ± s.e.m. for two independent experiments. See Figure 2—figure supplement 1E for Kd values associated with panel C. DOI:
http://dx.doi.org/10.7554/eLife.17101.006
Figure 2—figure supplement 1.. Quantifying the interaction of full-length UHRF1 and various mutants with histone H3 peptides and DNA oligonucleotides.
(A) FP binding assays quantifying the interaction of MBP-tagged UHRF1 H3mut with C-terminally FAM-labeled H31-20K9un and H31-20K9me3 peptides. Error is represented as ± s.e.m. for two independent experiments. (B) FP binding assays quantifying the interaction of UHRF1 with FAM-labeled HeDNA and UnDNA in the presence or absence of 10 μM unlabeled H31-20K9me3. Error is represented as ± s.e.m. for two independent experiments. (C) Table of the calculated Kd values for UHRF1 and MBP-UHRF1 binding to HeDNA or UnDNA in the presence or absence of unlabeled H31-20K9me3 (see also Figure 2C). The DNA and peptide binding activity of these constructs were similar; however, MBP-tagged protein displayed better solubility at higher protein concentration. (D) FP binding assays monitoring the interaction of Linkermut with HeDNA or UnDNA in the presence or absence of 10 μM unlabeled H31-20K9me3. Error is represented as ± s.e.m. for two independent experiments (E) Table of calculated Kd values for the indicated wild-type and mutant MBP-UHRF1 proteins binding to HeDNA or UnDNA in the presence or absence of unlabeled H31-20K9me3 (see also Figure 2C). DOI:
http://dx.doi.org/10.7554/eLife.17101.007
Figure 3.. DNA binding disrupts a UHRF1 intramolecular interaction.
(A) In vitro pull-down analysis of the interaction between GST-TTD-PHD and MBP or the indicated MBP fusions of UHRF1. (B) Pull-down analysis of the interaction between GST-TTD-PHD and MBP-SRA-RING (wild-type or DNAmut) fusions of UHRF1 in the presence or absence of the indicated DNA oligonucleotides. GST Ctrl is a GST fusion of the PHD-Bromo from BPTF (see Materials and methods). (C) Pull-down in the presence of H31-20K9me2. (D) Analytical size exclusion chromatography of UHRF1 in the absence or presence of HeDNA and H31-15K9me2. The calculated molecular weights for apo and ligand-bound UHRF1 are in agreement with the expected molecular weight of monomeric UHRF1. (E) Dynamic light scattering of UHRF1 in the absence or presence of HeDNA and H31-15K9me2. UHRF1 remains mono-dispersed (poly-dispersity < 25%) both in the presence and absence of the indicated ligands. The calculated mass range of 91–98 kD is in agreement with the expected molecular weight for full-length monomeric UHRF1, 90 kD. (F) Atomic force microscopy histograms of the volumes for 617 apo UHRF1 particles (left) and 884 HeDNA-bound UHRF1 particles (right). Distributions were fit to a single Gaussian peak using the peak fit function in Origin 6.1. DOI:
http://dx.doi.org/10.7554/eLife.17101.008
Figure 4.. UHRF1-mediated histone H3 ubiquitylation is stimulated by substrate and HeDNA recognition.
(A) UHRF1 ubiquitylation assays on an H31-32K9me2 peptide in the absence or presence of the indicated DNA oligonucleotides: HeDNA was titrated at semi-log intervals spanning 30 μM to 1 nM. SyDNA or UnDNA was added at 30 μM or 100 μM, respectively. (B) Rate measurement quantifying UHRF1 auto-ubiquitylation and H31-32K9me2 ubiquitylation in the presence of HeDNA or UnDNA at the indicated time points. Rate experiments were performed three times with similar results, and a representative blot is depicted. Blots were quantified using ImageQuant TL (GE Lifesciences). Quantified data was best described by a linear fit over the measured time scale, with the exception of HeDNA-stimulated H31-32K9me2 mono-ubiuitylation, which remained linear within the first 5 min of the reaction. (C) UHRF1 ubiquitylation assays on HeLa mononucleosomes in the presence of the indicated concentrations of HeDNA and/or an H31-15K9me2 peptide. (D) Ubiquitylation of an H31-43K9un peptide by UHRF1 and the indicated mutants (see Figure 1A for mutant annotation) in the absence or presence of HeDNA. DOI:
http://dx.doi.org/10.7554/eLife.17101.009
Figure 4—figure supplement 1.. UHRF1 ubiquitin ligase assays.
(A) Time course assay of UHRF1 auto-ubiquitylation in the presence of UnDNA or HeDNA in the absence of a histone peptide substrate (left). Rates of UHRF1 auto-ubiquitylation (right) were quantified from blots using ImageQuant TL (GE Lifesciences). Quantified data was best described by a linear fit over the measured time scale. (B) UHRF1 ubiquitylation of HeLa mononucleosomes in the presence or absence of HeDNA reveals that mononucleosome ubiquitylation is significantly enhanced in the presence of HeDNA. (C) Ubiquitylation of H31-32K9me2 by MBP-tagged UHRF1 and the indicated mutants. Mutant UHRF1 proteins display significant defects in HeDNA stimulated ubiquitylation activity with the exception of the D469G. (D) FP binding assays quantifying the interaction of the MBP-UHRF1 D469G with FAM-labeled HeDNA, SyDNA, or UnDNA. Error is represented as ± s.e.m. for two independent experiments. While the literature suggests a HeDNA binding defect for the D469G mutation, these assays reveal this mutant binds DNA similarly to wild-type (for comparison see Figure 1B). DOI:
http://dx.doi.org/10.7554/eLife.17101.010
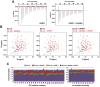
Figure 5.. HeDNA binding directs ubiquitin to histone substrates.
(A) ITC measuring the interaction of UHRF1 with E2-N-ub (UbcH5c(C85K)-ub linked by isopeptide bond) in the presence and absence of HeDNA (see also Figure 5—figure supplement 1A). (B) Average peak intensities for 1H-15N HSQC-TROSY spectra of the 15N-E2-o-ub (E2-o-Ub, UbcH5c(S22R/C85S)-ub esterified conjugate) (see also Figure 5—figure supplement 1B–C). Percentages indicate the reduction in intensity due to addition of UHRF1 or HeDNA. The addition of HeDNA to E2-ub (comparing blue to red) results in the same decrease in intensity as the addition of HeDNA to a sample containing E2-ub and UHRF1 (comparing green to purple) for both the E2 or ub within the conjugate. (C) Coomassie-stained gel of ubiquitin discharge assays in the presence of the indicated ligands and 20 mM free lysine (left). Densitometry analysis of the indicated components of the reaction (right). Line coloring corresponds to lane labels at the top of the gel. (D) Coomassie-stained gel of ubiquitin discharge assays in the presence HeDNA and either no peptide, H31-20, H31-20K14aK18ac, H31-20K9acK14acK18ac and 20 mM free lysine (left). Densitometry analysis of the indicated components of the reaction (right). Line coloring corresponds to lane labels at the top of the gel. We conducted at least five ubiquitin discharge assays, and the trends observed for each condition in panels C and D were consistent across all experiments. DOI:
http://dx.doi.org/10.7554/eLife.17101.011
Figure 5—figure supplement 1.. HeDNA binding does not modulate the interaction of UHRF1 with E2-ubiquitin conjugate.
(A) Isotherm from ITC experiments monitoring E2-N-Ub binding to UHRF1 in the absence or presence of HeDNA. (B), NMR analysis of UHRF1 interaction with E2-conjugated ubiquitin in the absence or presence of HeDNA. 1H-15N HSQC-TROSY spectra of the 15N-E2-o-ub conjugate (UbcH5c(Ser22Arg/Cys85Ser-O-ub; 200 μM) in the presence or absence of HeDNA (0.11 Molar equivalents) and/or MBP-UHRF1 (0.09 Molar equivalents). HeDNA does not appear to significantly perturb the 15N-E2-o-ub spectrum (left). MBP-UHRF1 binds to the E2-ub conjugate as indicated by peak intensity loss (middle), however HeDNA does not promote further binding (right). (C) NMR peak intensities relative to the 15N-E2-o-ub conjugate spectrum for the E2 UbcH5c (left) and ub (right). DOI:
http://dx.doi.org/10.7554/eLife.17101.012
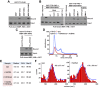
Figure 6.. HeDNA stimulates UHRF1-directed ubiquitylation of multiple N-terminal lysines on histone H3.
(A) Schematic of the assay and sample preparation strategy to identify by LC-MC/MS products of UHRF1 ubiquitylation reactions with HeLa mononucleosomes in the presence of UnDNA or HeDNA. (B) Quantification of the area under the curve (AUC) from extracted-ion chromatograms for the indicated ubiquitylated H3 peptides enriched by immunoprecipitation of FLAG-ub. See Figure 6—figure supplement 1 for retention times and fragmentation for identified peptides. (C) Immunoblot analysis for Flag-ub and the indicated histone PTMs following UHRF1 ubiquitylation of HeLa mononucleosomes reacted in the presence of HeDNA or UnDNA, (-) indicates unreacted nucleosomes. DOI:
http://dx.doi.org/10.7554/eLife.17101.013
Figure 6—figure supplement 1.. Ion-extracted chromatograms (left) and fragmentation patterns (right) for each ubiquitylated peptide identified using the search procedures described in Materials and methods.
DOI:
http://dx.doi.org/10.7554/eLife.17101.014
Figure 6—figure supplement 2.. Characterizing lysine prioritization of UHRF1 ubiquitylation on mononucleosomes.
(A) Ubiquitylation of Hela mononucleosomes after 2 hr in the presence of HeDNA or UnDNA. (B) Ratio and normalized ratio (HeDNA/UnDNA) for H3 ubiquitylated peptides from propionylated samples. Samples were normalized to the mean ratio of free ubiquitin peptides. Quantification was performed with Skyline and values for each peptide charge state were summed. (C) Ratio and normalized ratio (HeDNA/UnDNA) for H3 ubiquitylated peptides from propionylated and unreacted samples. A ubiquitylated peptide from the PHD (marked with an asterisk) was enriched in the HeDNA sample. (D) Location of UHRF1 auto-ubiquitylation sites lining the TTD-PHD. TTD, pink; PHD, blue; modified lysines, red. The C-terminal atom on the H3 peptide (S10) is shown as a sphere. (E) Auto-ubiquitylation of UHRF1 in the absence of H3 peptides with either HeDNA or UnDNA when run under conditions to resolve higher molecular weight species. DOI:
http://dx.doi.org/10.7554/eLife.17101.015
Figure 6—figure supplement 3.. UHRF1 targets H3K9me2 histones for ubiquitylation.
HeDNA-stimulated UHRF1 ubiquitylation assays titrating recombinant histones H2A, H2B, H3, or H3K9me2 (22.5 μM, 7.5 μM, 2.5 μM, 0.75 μM, 0.225 μM, 0.075 μM, 0.025 μM). DOI:
http://dx.doi.org/10.7554/eLife.17101.016
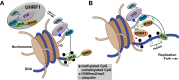
Figure 7.. Proposed model for the contributions of DNA and histone recognition events to the DNA methylation regulatory function of UHRF1.
(A) UHRF1 is targeted to and retained on chromatin by the combined actions of H3K9me2/me3 recognition through the TTD-PHD and DNA interaction, independent of methylation status, through the SRA. (B) The interaction of the SRA with HeDNA, a DNA replication intermediate, directs the ubiquitin ligase activity of UHRF1 towards N-terminal lysines on the histone H3 tail. H3 ubiquitylation by UHRF1 contributes to the retention of DNMT1 in chromatin environments enriched for HeDNA and facilitates the epigenetic inheritance of DNA methylation patterns. DOI:
http://dx.doi.org/10.7554/eLife.17101.017
Figure 7—figure supplement 1.. Mouse UHRF1 (Np95) SRA adopts different conformations bound to UnDNA (left;PDB:2ZO2) and HeDNA (right;PDB:3F8I).
DOI:
http://dx.doi.org/10.7554/eLife.17101.018
Similar articles
- A Bifunctional Role for the UHRF1 UBL Domain in the Control of Hemi-methylated DNA-Dependent Histone Ubiquitylation.
DaRosa PA, Harrison JS, Zelter A, Davis TN, Brzovic P, Kuhlman B, Klevit RE. DaRosa PA, et al. Mol Cell. 2018 Nov 15;72(4):753-765.e6. doi: 10.1016/j.molcel.2018.09.029. Epub 2018 Nov 1. Mol Cell. 2018. PMID: 30392931 Free PMC article. - Critical Role of the UBL Domain in Stimulating the E3 Ubiquitin Ligase Activity of UHRF1 toward Chromatin.
Foster BM, Stolz P, Mulholland CB, Montoya A, Kramer H, Bultmann S, Bartke T. Foster BM, et al. Mol Cell. 2018 Nov 15;72(4):739-752.e9. doi: 10.1016/j.molcel.2018.09.028. Epub 2018 Nov 1. Mol Cell. 2018. PMID: 30392929 Free PMC article. - Chromatin structure and its chemical modifications regulate the ubiquitin ligase substrate selectivity of UHRF1.
Vaughan RM, Dickson BM, Whelihan MF, Johnstone AL, Cornett EM, Cheek MA, Ausherman CA, Cowles MW, Sun ZW, Rothbart SB. Vaughan RM, et al. Proc Natl Acad Sci U S A. 2018 Aug 28;115(35):8775-8780. doi: 10.1073/pnas.1806373115. Epub 2018 Aug 13. Proc Natl Acad Sci U S A. 2018. PMID: 30104358 Free PMC article. - Regulation of maintenance DNA methylation via histone ubiquitylation.
Nishiyama A, Yamaguchi L, Nakanishi M. Nishiyama A, et al. J Biochem. 2016 Jan;159(1):9-15. doi: 10.1093/jb/mvv113. Epub 2015 Nov 20. J Biochem. 2016. PMID: 26590302 Free PMC article. Review. - Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.
Bronner C, Alhosin M, Hamiche A, Mousli M. Bronner C, et al. Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
Cited by
- The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.
Wapenaar H, Clifford G, Rolls W, Pasquier M, Burdett H, Zhang Y, Deák G, Zou J, Spanos C, Taylor MRD, Mills J, Watson JA, Kumar D, Clark R, Das A, Valsakumar D, Bramham J, Voigt P, Sproul D, Wilson MD. Wapenaar H, et al. EMBO Rep. 2024 Nov 11. doi: 10.1038/s44319-024-00306-3. Online ahead of print. EMBO Rep. 2024. PMID: 39528729 - Design of linked-domain protein inhibitors of UBE2D as tools to study cellular ubiquitination.
Bukhari Z, Gu L, Nederstigt AE, Cope LJ, Bolhuis DL, Harvey K, Allen T, Hill S, Yang Y, Lawson G, Lu C, Tran T, Pineda L, Low L, Chiang A, Song J, Fong MV, Rangel VM, Chan WK, Kleiger G, Goldfarb D, Vierra CA, Brown NG, Harrison JS. Bukhari Z, et al. bioRxiv [Preprint]. 2024 Sep 2:2024.09.02.610852. doi: 10.1101/2024.09.02.610852. bioRxiv. 2024. PMID: 39282319 Free PMC article. Preprint. - CDCA7 is an evolutionarily conserved hemimethylated DNA sensor in eukaryotes.
Wassing IE, Nishiyama A, Shikimachi R, Jia Q, Kikuchi A, Hiruta M, Sugimura K, Hong X, Chiba Y, Peng J, Jenness C, Nakanishi M, Zhao L, Arita K, Funabiki H. Wassing IE, et al. Sci Adv. 2024 Aug 23;10(34):eadp5753. doi: 10.1126/sciadv.adp5753. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178260 Free PMC article. - Oncogenic Roles of UHRF1 in Cancer.
Kim A, Benavente CA. Kim A, et al. Epigenomes. 2024 Jul 1;8(3):26. doi: 10.3390/epigenomes8030026. Epigenomes. 2024. PMID: 39051184 Free PMC article. Review. - The evolutionary consequences of interactions between the epigenome, the genome and the environment.
Baduel P, Sammarco I, Barrett R, Coronado-Zamora M, Crespel A, Díez-Rodríguez B, Fox J, Galanti D, González J, Jueterbock A, Wootton E, Harney E. Baduel P, et al. Evol Appl. 2024 Jul 23;17(7):e13730. doi: 10.1111/eva.13730. eCollection 2024 Jul. Evol Appl. 2024. PMID: 39050763 Free PMC article. Review.
References
- Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M, Shirakawa M. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. PNAS. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R00 CA181343/CA/NCI NIH HHS/United States
- R01 GM110058/GM/NIGMS NIH HHS/United States
- T32 CA009156/CA/NCI NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- K99 CA181343/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 GM088055/GM/NIGMS NIH HHS/United States
- R01 GM073960/GM/NIGMS NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R01 GM079480/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources