MreB Orientation Correlates with Cell Diameter in Escherichia coli - PubMed (original) (raw)
MreB Orientation Correlates with Cell Diameter in Escherichia coli
Nikolay Ouzounov et al. Biophys J. 2016.
Abstract
Bacteria have remarkably robust cell shape control mechanisms. For example, cell diameter only varies by a few percent across a given population. The bacterial actin homolog, MreB, is necessary for establishment and maintenance of rod shape although the detailed properties of MreB that are important for shape control remained unknown. In this study, we perturb MreB in two ways: by treating cells with the polymerization-inhibiting drug A22 and by creating point mutants in mreB. These perturbations modify the steady-state diameter of cells over a wide range, from 790 ± 30 nm to 1700 ± 20 nm. To determine which properties of MreB are important for diameter control, we correlated structural characteristics of fluorescently tagged MreB polymers with cell diameter by simultaneously analyzing three-dimensional images of MreB and cell shape. Our results indicate that the helical pitch angle of MreB inversely correlates with the cell diameter of Escherichia coli. Other correlations between MreB and cell diameter are not found to be significant. These results demonstrate that the physical properties of MreB filaments are important for shape control and support a model in which MreB organizes the cell wall growth machinery to produce a chiral cell wall structure and dictate cell diameter.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
Cell shape and polymer fitting method. (A) Diagram outlines the cell shape and polymer fitting algorithm. Cells expressing MreBmsfGFP under native regulation are membrane-stained with FM4-64 and imaged using 3D fluorescent microscopy. The imaging process can be written as a convolution between the point-spread function (PSF) of our microscope and the spatial distribution of fluorescent molecules, in this case the membrane. To estimate the shape of the surface, a model cell is convolved with the PSF to create a 3D simulated image. The surface is relaxed so that the simulated image best matches the experimental image. A similar process is used to fit the MreB polymers. Each polymer is modeled as a stiff elastic rod confined to the surface of the membrane. Again, a simulated 3D MreB image is created and model filaments relax to best match the experimental image. (B) Representative surface fits of cells expressing MreBmsfGFP are shown. The color of the surface is determined by interpolating the intensity of the 3D MreB image at the points of the surface. The detected polymers are shown in black. To see this figure in color, go online.
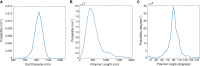
Figure 2
Probability density functions of (A) cell diameter, (B) MreB polymer lengths, and (C) MreB monomer angles in E. coli expressing MreBmsfGFP. Data is collected from 459 cells, with an average of 7.3 polymers detected per cell. The distribution in (A) shows an average diameter of 934 ± 6 nm. The average MreB polymer length was measured as 500 ± 10 nm. The angle distribution of monomers in (C) is made by weighting the angle distribution of polymers by the length of each polymer, and had a mean angle of 91 ± 1°. All values are shown as mean ± 80% confidence intervals. To see this figure in color, go online.
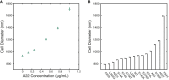
Figure 3
Two independent methods to perturb cell diameter. (A) Cells expressing MreBmsfGFP were grown to steady state in different concentrations of the polymerization-inhibiting drug A22. All treatments are below the lethal concentrations of A22. As A22 concentration increases, cells significantly increase their diameter. (B) Cell diameter can also be changed with single-point mutants in the mreB msfGFP. Fourteen mutants were generated with diameters both larger and smaller than the unmutated form (WT). Mutants are arranged in order of increasing diameter. Error bars represent 80% confidence intervals. To see this figure in color, go online.
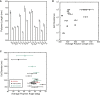
Figure 4
MreB polymer measurements. (A) MreB mutations can alter the MreB polymer length. In some cases, the polymer length is increased with respect of wild-type and in other cases the polymer length is decreased. Mutations are arranged as in Fig. 3_B_, in order of increasing average cell diameter. Error bars represent 80% confidence intervals. (B) MreB polymer length appears to correlate with the level of A22 resistance of the MreB mutants. Mutations that have longer polymers have high levels of A22 resistance whereas increased sensitivity to A22 is seen in mutations that have shorter polymers. A22 resistance levels are shown as a fold changes from wild-type MreBmsfGFP, with levels capped at 100× that of wild-type since higher levels of A22 can effect proteins other than MreB. (C) The average polymer angle inversely correlates with the cell diameter in both mreB mutant data (black) and A22 treatment data (green). The wild-type mreB is shown in red. The cell diameter increases as the average helical pitch angle of MreB decreases. The handedness of the helical pitch changes in both data sets as 90° is crossed. To see this figure in color, go online.
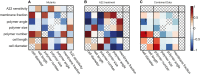
Figure 5
Cell shape was modified using either point mutants in mreB msfGFP or treatment with sublethal concentrations of A22. Maps of Pearson correlation coefficients between cell shape metrics and measured MreB properties were created for each set of conditions, for the (A) mutants (15 conditions), (B) A22 treatment (6 conditions), and (C) combined data sets (19 conditions). In (C), † indicates p = .023 and ‡ indicates p < .001, after accounting for multiple comparison. To see this figure in color, go online.
Similar articles
- RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis.
Morgenstein RM, Bratton BP, Nguyen JP, Ouzounov N, Shaevitz JW, Gitai Z. Morgenstein RM, et al. Proc Natl Acad Sci U S A. 2015 Oct 6;112(40):12510-5. doi: 10.1073/pnas.1509610112. Epub 2015 Sep 22. Proc Natl Acad Sci U S A. 2015. PMID: 26396257 Free PMC article. - In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2.
Varma A, Young KD. Varma A, et al. J Bacteriol. 2009 Jun;191(11):3526-33. doi: 10.1128/JB.01812-08. Epub 2009 Apr 3. J Bacteriol. 2009. PMID: 19346310 Free PMC article. - The bacterial actin MreB rotates, and rotation depends on cell-wall assembly.
van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. van Teeffelen S, et al. Proc Natl Acad Sci U S A. 2011 Sep 20;108(38):15822-7. doi: 10.1073/pnas.1108999108. Epub 2011 Sep 8. Proc Natl Acad Sci U S A. 2011. PMID: 21903929 Free PMC article. - How to Build a Bacterial Cell: MreB as the Foreman of E. coli Construction.
Shi H, Bratton BP, Gitai Z, Huang KC. Shi H, et al. Cell. 2018 Mar 8;172(6):1294-1305. doi: 10.1016/j.cell.2018.02.050. Cell. 2018. PMID: 29522748 Free PMC article. Review. - Polar localization of MreB actin is inhibited by anionic phospholipids in the rod-shaped bacterium Escherichia coli.
Shiomi D. Shiomi D. Curr Genet. 2017 Oct;63(5):845-848. doi: 10.1007/s00294-017-0696-5. Epub 2017 Apr 24. Curr Genet. 2017. PMID: 28439631 Review.
Cited by
- Emergence of Escherichia coli critically buckled motile helices under stress.
Phan TV, Morris RJ, Lam HT, Hulamm P, Black ME, Bos J, Austin RH. Phan TV, et al. Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):12979-12984. doi: 10.1073/pnas.1809374115. Epub 2018 Nov 29. Proc Natl Acad Sci U S A. 2018. PMID: 30498027 Free PMC article. - MreB Forms Subdiffraction Nanofilaments during Active Growth in Bacillus subtilis.
Billaudeau C, Yao Z, Cornilleau C, Carballido-López R, Chastanet A. Billaudeau C, et al. mBio. 2019 Jan 29;10(1):e01879-18. doi: 10.1128/mBio.01879-18. mBio. 2019. PMID: 30696741 Free PMC article. - Chiral twisting in a bacterial cytoskeletal polymer affects filament size and orientation.
Shi H, Quint DA, Grason GM, Gopinathan A, Huang KC. Shi H, et al. Nat Commun. 2020 Mar 16;11(1):1408. doi: 10.1038/s41467-020-14752-9. Nat Commun. 2020. PMID: 32179732 Free PMC article. - CRISPR interference to interrogate genes that control biofilm formation in Pseudomonas fluorescens.
Noirot-Gros MF, Forrester S, Malato G, Larsen PE, Noirot P. Noirot-Gros MF, et al. Sci Rep. 2019 Nov 4;9(1):15954. doi: 10.1038/s41598-019-52400-5. Sci Rep. 2019. PMID: 31685917 Free PMC article. - Cardiolipin Alters Rhodobacter sphaeroides Cell Shape by Affecting Peptidoglycan Precursor Biosynthesis.
Lin TY, Gross WS, Auer GK, Weibel DB. Lin TY, et al. mBio. 2019 Feb 19;10(1):e02401-18. doi: 10.1128/mBio.02401-18. mBio. 2019. PMID: 30782656 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases