Rapid Evolutionary Rates and Unique Genomic Signatures Discovered in the First Reference Genome for the Southern Ocean Salp, Salpa thompsoni (Urochordata, Thaliacea) - PubMed (original) (raw)
Rapid Evolutionary Rates and Unique Genomic Signatures Discovered in the First Reference Genome for the Southern Ocean Salp, Salpa thompsoni (Urochordata, Thaliacea)
Nathaniel K Jue et al. Genome Biol Evol. 2016.
Abstract
A preliminary genome sequence has been assembled for the Southern Ocean salp, Salpa thompsoni (Urochordata, Thaliacea). Despite the ecological importance of this species in Antarctic pelagic food webs and its potential role as an indicator of changing Southern Ocean ecosystems in response to climate change, no genomic resources are available for S. thompsoni or any closely related urochordate species. Using a multiple-platform, multiple-individual approach, we have produced a 318,767,936-bp genome sequence, covering >50% of the estimated 602 Mb (±173 Mb) genome size for S. thompsoni Using a nonredundant set of predicted proteins, >50% (16,823) of sequences showed significant homology to known proteins and ∼38% (12,151) of the total protein predictions were associated with Gene Ontology functional information. We have generated 109,958 SNP variant and 9,782 indel predictions for this species, serving as a resource for future phylogenomic and population genetic studies. Comparing the salp genome to available assemblies for four other urochordates, Botryllus schlosseri, Ciona intestinalis, Ciona savignyi and Oikopleura dioica, we found that S. thompsoni shares the previously estimated rapid rates of evolution for these species. High mutation rates are thus independent of genome size, suggesting that rates of evolution >1.5 times that observed for vertebrates are a broad taxonomic characteristic of urochordates. Tests for positive selection implemented in PAML revealed a small number of genes with sites undergoing rapid evolution, including genes involved in ribosome biogenesis and metabolic and immune process that may be reflective of both adaptation to polar, planktonic environments as well as the complex life history of the salps. Finally, we performed an initial survey of small RNAs, revealing the presence of known, conserved miRNAs, as well as novel miRNA genes; unique piRNAs; and mature miRNA signatures for varying developmental stages. Collectively, these resources provide a genomic foundation supporting S. thompsoni as a model species for further examination of the exceptional rates and patterns of genomic evolution shown by urochordates. Additionally, genomic data will allow for the development of molecular indicators of key life history events and processes and afford new understandings and predictions of impacts of climate change on this key species of Antarctic pelagic ecosystems.
Keywords: Antarctic ecosystem; Salpa thompsoni; Thaliacean genome; miRNA; urochordate.
© The Author(s) 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Fig. 1.—
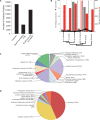
(A) Evolutionary position of Salpa thompsoni with respect to other groups within the Subphylum Tunicata, including Ciona spp. and Botryllus schlosseri within Class Ascidiacea, Oikopleura dioica within Class Appendicularia, as well as Subphyla Cephalochrodata and Vertebrata. Overall relationships among Eumetazoa are indicated to left. (B) Map of Antarctica indicating the collection area for S. thompsoni samples (boxed inset, red diamond) in the Southern Indian Ocean. (C) Reproductive life cycle of S. thompsoni.
Fig. 2.—
Salpa thompsoni gene annotations. (A) Annotation status of S. thompsoni predicted proteins delineated by validation method. (B) The number of ambiguous sequences, codon shifts (left) and total orthologues (right) with S. thompsoni are shown for each species (bottom). Phylogenetic relationships for each species are indicated. (C and D) Pie chart summary of level 2 gene ontology terms associated with biological process (C) and molecular function (D) categories for all annotated Annotation status of S. thompsoni predicted proteins.
Fig. 3.—
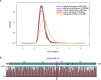
(A) Kernel density plots of ortholog sequence divergence from Cephalochordate Branchiostoma floridae for each Urchordate species (see legend) reveal similar evolutionary rates among Urochordate species, with the exception of Oikopleura dioica which appears to evolve more rapidly than the others. Orthology groups (n = 967) investigated for positive selection were used in this analysis. (B) Schematic of cyclophilin A protein sequence with percent sequence conservation at each site shown for other Urochordate and Cephalochorate species analyzed. Yellow box indicates site under positive selection; purple boxes indicate metal ion-binding site; blue arrow indicates propyl isomerase functional domain.
Fig. 4.—
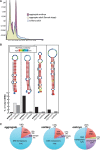
(A) Size distribution of small RNAs post-trimming across embryo and adult solitary salp and adult female samples. (B) miRNA precursor fold predictions for each novel miRNA. Nucleotide is color coded by probability, the mature miRNA is indicated by a black line for each hairpin, and sample distribution for each is indicated. (C) Classifiable piRNA content for aggregate female stage, solitary adult and embryo samples, excluding tRNA, rRNA and unmasked (nonrepeat) sequences within the piRNA size range.
Similar articles
- Gene expression patterns of Salpa thompsoni reveal remarkable differences in metabolism and reproduction near the Antarctic Polar Front.
Müller SJ, Pakhomov EA, Urso I, Sales G, Pittà C, Michael K, Meyer B. Müller SJ, et al. Biol Lett. 2023 Dec;19(12):20230274. doi: 10.1098/rsbl.2023.0274. Epub 2023 Dec 6. Biol Lett. 2023. PMID: 38053363 Free PMC article. - Salpa genome and developmental transcriptome analyses reveal molecular flexibility enabling reproductive success in a rapidly changing environment.
Castellano KR, Batta-Lona P, Bucklin A, O'Neill RJ. Castellano KR, et al. Sci Rep. 2023 Nov 29;13(1):21056. doi: 10.1038/s41598-023-47429-6. Sci Rep. 2023. PMID: 38030690 Free PMC article. - Altered miRNA repertoire in the simplified chordate, Oikopleura dioica.
Fu X, Adamski M, Thompson EM. Fu X, et al. Mol Biol Evol. 2008 Jun;25(6):1067-80. doi: 10.1093/molbev/msn060. Epub 2008 Mar 13. Mol Biol Evol. 2008. PMID: 18339653 - Urochordate genomes.
Satoh N, Kawashima T, Shoguchi E, Satou Y. Satoh N, et al. Genome Dyn. 2006;2:198-212. doi: 10.1159/000095105. Genome Dyn. 2006. PMID: 18753780 Review. - An advanced filter-feeder hypothesis for urochordate evolution.
Satoh N. Satoh N. Zoolog Sci. 2009 Feb;26(2):97-111. doi: 10.2108/zsj.26.97. Zoolog Sci. 2009. PMID: 19341326 Review.
Cited by
- De novo genome assembly and comparative genomics for the colonial ascidian Botrylloides violaceus.
Sumner JT, Andrasz CL, Johnson CA, Wax S, Anderson P, Keeling EL, Davidson JM. Sumner JT, et al. G3 (Bethesda). 2023 Sep 30;13(10):jkad181. doi: 10.1093/g3journal/jkad181. G3 (Bethesda). 2023. PMID: 37555394 Free PMC article. - A phylogenomic framework and timescale for comparative studies of tunicates.
Delsuc F, Philippe H, Tsagkogeorga G, Simion P, Tilak MK, Turon X, López-Legentil S, Piette J, Lemaire P, Douzery EJP. Delsuc F, et al. BMC Biol. 2018 Apr 13;16(1):39. doi: 10.1186/s12915-018-0499-2. BMC Biol. 2018. PMID: 29653534 Free PMC article. - nrDNA:mtDNA copy number ratios as a comparative metric for evolutionary and conservation genetics.
Goodall-Copestake WP. Goodall-Copestake WP. Heredity (Edinb). 2018 Aug;121(2):105-111. doi: 10.1038/s41437-018-0088-8. Epub 2018 May 12. Heredity (Edinb). 2018. PMID: 29752470 Free PMC article. - ANISEED 2017: extending the integrated ascidian database to the exploration and evolutionary comparison of genome-scale datasets.
Brozovic M, Dantec C, Dardaillon J, Dauga D, Faure E, Gineste M, Louis A, Naville M, Nitta KR, Piette J, Reeves W, Scornavacca C, Simion P, Vincentelli R, Bellec M, Aicha SB, Fagotto M, Guéroult-Bellone M, Haeussler M, Jacox E, Lowe EK, Mendez M, Roberge A, Stolfi A, Yokomori R, Brown CT, Cambillau C, Christiaen L, Delsuc F, Douzery E, Dumollard R, Kusakabe T, Nakai K, Nishida H, Satou Y, Swalla B, Veeman M, Volff JN, Lemaire P. Brozovic M, et al. Nucleic Acids Res. 2018 Jan 4;46(D1):D718-D725. doi: 10.1093/nar/gkx1108. Nucleic Acids Res. 2018. PMID: 29149270 Free PMC article. - An in situ digital synthesis strategy for the discovery and description of ocean life.
Burns JA, Becker KP, Casagrande D, Daniels J, Roberts P, Orenstein E, Vogt DM, Teoh ZE, Wood R, Yin AH, Genot B, Gruber DF, Katija K, Wood RJ, Phillips BT. Burns JA, et al. Sci Adv. 2024 Jan 19;10(3):eadj4960. doi: 10.1126/sciadv.adj4960. Epub 2024 Jan 17. Sci Adv. 2024. PMID: 38232174 Free PMC article.
References
- Abrusán G, Grundmann N, DeMester L, Makalowski W. 2009. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25:1329–1330. - PubMed
- Alldredge A, Madin L. 1982. Pelagic tunicates: unique herbivores in the marine plankton. Bioscience 32:655–663.
- Atkinson A, Siegel V, Pakhomov E, Rothery P. 2004. Long-term decline in krill stock and increase in salps within the Southern Ocean. Nature 432:100–103. - PubMed
- Batta-Lona PG, Maas AE, O’Neill RJ, Wiebe PH, Bucklin A. 2016. (submitted). Transcriptome-wide profiles of gene expression of Salpa thompsoni in relation to variation of the pelagic environment of the Southern Ocean. Polar Biol.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous