DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering - PubMed (original) (raw)
DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering
Daniel Franke et al. J Appl Crystallogr. 2009.
Abstract
DAMMIF, a revised implementation of the _ab_-initio shape-determination program DAMMIN for small-angle scattering data, is presented. The program was fully rewritten, and its algorithm was optimized for speed of execution and modified to avoid limitations due to the finite search volume. Symmetry and anisometry constraints can be imposed on the particle shape, similar to DAMMIN. In equivalent conditions, DAMMIF is 25-40 times faster than DAMMIN on a single CPU. The possibility to utilize multiple CPUs is added to DAMMIF. The application is available in binary form for major platforms.
Keywords: DAMMIF; DAMMIN; computer programs; particle shape determination; small-angle scattering.
Figures
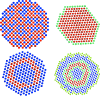
Figure 1
Cross sections of dummy atom models of DAMMIN (left) and DAMMIF (right). The top row shows initial models (randomized and proto-particle) and the bottom row the final models by the two programs. The different colours indicate particle (red) and solvent (turquoise, blue and green) states of the dummy atoms. In DAMMIF, only red and turquoise beads are subject to phase changes; DAMMIN generally allows phase transitions anywhere in the search volume. Green solvent beads indicate the current, extensible, border of _DAMMIF_’s mapped area (all visible beads).
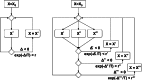
Figure 2
SA algorithm as implemented in DAMMIN (left) and DAMMIF (right). An initial starting model  is refined to yield the best possible fit to the experimental data. In DAMMIN, only one neighbouring model
is refined to yield the best possible fit to the experimental data. In DAMMIN, only one neighbouring model  is taken into account at a time. If multiple cores or CPUs are available, it is possible to prefetch multiple models in parallel, here shown as
is taken into account at a time. If multiple cores or CPUs are available, it is possible to prefetch multiple models in parallel, here shown as  ,
,  ,
,  . Each prefetched model is then examined and either accepted or rejected, according to the rules of SA.
. Each prefetched model is then examined and either accepted or rejected, according to the rules of SA.
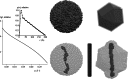
Figure 3
Reconstruction of a cylindrical particle with radius 10 Å and height 200 Å (bottom centre) from its simulated scattering pattern presented on the left-hand side [relative intensity I versus inverse ångströms; the distance distribution function p(r) computed by GNOM is displayed in the insert]. The starting (top row) and final (bottom row) models from DAMMIN and DAMMIF are displayed in the middle and right panels, respectively. DAMMIN ran in a slow mode inside the spherical search volume (packing radius  Å, CPU time used 246 min). For DAMMIF, the value of
Å, CPU time used 246 min). For DAMMIF, the value of  was 3.0 Å and the run on the same single processor took 8 min.
was 3.0 Å and the run on the same single processor took 8 min.
Similar articles
- Rapid shape determination of tissue transglutaminase using high-throughput computing.
Lammie D, Osborne J, Aeschlimann D, Wess TJ. Lammie D, et al. Acta Crystallogr D Biol Crystallogr. 2007 Sep;63(Pt 9):1022-4. doi: 10.1107/S0907444907032933. Epub 2007 Aug 17. Acta Crystallogr D Biol Crystallogr. 2007. PMID: 17704572 - Ambiguity assessment of small-angle scattering curves from monodisperse systems.
Petoukhov MV, Svergun DI. Petoukhov MV, et al. Acta Crystallogr D Biol Crystallogr. 2015 May;71(Pt 5):1051-8. doi: 10.1107/S1399004715002576. Epub 2015 Apr 24. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25945570 - Direct shape determination of intermediates in evolving macromolecular solutions from small-angle scattering data.
Konarev PV, Svergun DI. Konarev PV, et al. IUCrJ. 2018 May 30;5(Pt 4):402-409. doi: 10.1107/S2052252518005900. eCollection 2018 Jul 1. IUCrJ. 2018. PMID: 30002841 Free PMC article. - Application of SAXS for the Structural Characterization of IDPs.
Kachala M, Valentini E, Svergun DI. Kachala M, et al. Adv Exp Med Biol. 2015;870:261-89. doi: 10.1007/978-3-319-20164-1_8. Adv Exp Med Biol. 2015. PMID: 26387105 Review. - Advances in structure analysis using small-angle scattering in solution.
Svergun DI, Koch MH. Svergun DI, et al. Curr Opin Struct Biol. 2002 Oct;12(5):654-60. doi: 10.1016/s0959-440x(02)00363-9. Curr Opin Struct Biol. 2002. PMID: 12464319 Review.
Cited by
- Conformational changes in a hyperthermostable glycoside hydrolase: enzymatic activity is a consequence of the loop dynamics and protonation balance.
de Oliveira LC, da Silva VM, Colussi F, Cabral AD, de Oliveira Neto M, Squina FM, Garcia W. de Oliveira LC, et al. PLoS One. 2015 Feb 27;10(2):e0118225. doi: 10.1371/journal.pone.0118225. eCollection 2015. PLoS One. 2015. PMID: 25723179 Free PMC article. - Mechanism of efficient double-strand break repair by a long non-coding RNA.
Thapar R, Wang JL, Hammel M, Ye R, Liang K, Sun C, Hnizda A, Liang S, Maw SS, Lee L, Villarreal H, Forrester I, Fang S, Tsai MS, Blundell TL, Davis AJ, Lin C, Lees-Miller SP, Strick TR, Tainer JA. Thapar R, et al. Nucleic Acids Res. 2020 Nov 4;48(19):10953-10972. doi: 10.1093/nar/gkaa784. Nucleic Acids Res. 2020. PMID: 33045735 Free PMC article. - Role of Conserved Proline Residues in Human Apolipoprotein A-IV Structure and Function.
Deng X, Walker RG, Morris J, Davidson WS, Thompson TB. Deng X, et al. J Biol Chem. 2015 Apr 24;290(17):10689-702. doi: 10.1074/jbc.M115.637058. Epub 2015 Mar 2. J Biol Chem. 2015. PMID: 25733664 Free PMC article. - Thermodynamic coupling of the tandem RRM domains of hnRNP A1 underlie its pleiotropic RNA binding functions.
Levengood JD, Potoyan D, Penumutchu S, Kumar A, Zhou Q, Wang Y, Hansen AL, Kutluay S, Roche J, Tolbert BS. Levengood JD, et al. Sci Adv. 2024 Jul 12;10(28):eadk6580. doi: 10.1126/sciadv.adk6580. Epub 2024 Jul 10. Sci Adv. 2024. PMID: 38985864 Free PMC article. - Molecular basis of F-actin regulation and sarcomere assembly via myotilin.
Kostan J, Pavšič M, Puž V, Schwarz TC, Drepper F, Molt S, Graewert MA, Schreiner C, Sajko S, van der Ven PFM, Onipe A, Svergun DI, Warscheid B, Konrat R, Fürst DO, Lenarčič B, Djinović-Carugo K. Kostan J, et al. PLoS Biol. 2021 Apr 12;19(4):e3001148. doi: 10.1371/journal.pbio.3001148. eCollection 2021 Apr. PLoS Biol. 2021. PMID: 33844684 Free PMC article.
References
- Bentley, J. L. (1975). Commun. ACM, 18, 509–517.
- Dagum, L. & Menon, R. (1998). IEEE Comput. Sci. Eng. 5, 46–55.
- Debye, P. (1915). Ann. Phys. 46, 809–823.
LinkOut - more resources
Full Text Sources
Other Literature Sources