RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3 - PubMed (original) (raw)
RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3
Min Xie et al. Cancer Res. 2016.
Abstract
Despite advances in diagnosis and treatment, prostate cancer is the most prevalent cancer in males and the second highest cause of cancer-related mortality. We identified an RNA helicase gene, DDX3 (DDX3X), which is overexpressed in prostate cancers, and whose expression is directly correlated with high Gleason scores. Knockdown of DDX3 in the aggressive prostate cancer cell lines DU145 and 22Rv1 resulted in significantly reduced clonogenicity. To target DDX3, we rationally designed a small molecule, RK-33, which docks into the ATP-binding domain of DDX3. Functional studies indicated that RK-33 preferentially bound to DDX3 and perturbed its activity. RK-33 treatment of prostate cancer cell lines DU145, 22Rv1, and LNCaP (which have high DDX3 levels) decreased proliferation and induced a G1 phase cell-cycle arrest. Conversely, the low DDX3-expressing cell line, PC3, exhibited few changes following RK-33 treatment. Importantly, combination studies using RK-33 and radiation exhibited synergistic effects both in vitro and in a xenograft model of prostate cancer demonstrating the role of RK-33 as a radiosensitizer. Taken together, these results indicate that blocking DDX3 by RK-33 in combination with radiation treatment is a viable option for treating locally advanced prostate cancer. Cancer Res; 76(21); 6340-50. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
V. Raman, P.J. van Diest, and G.M. Bol have ownership interest (including patents) as a co-owner of a patent on DDX3 targeting. No potential conflicts of interest were disclosed by the other authors.
Figures
Figure 1
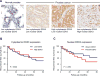
Correlation of DDX3 expression and prostate cancer. A, the staining intensity of DDX3 in normal (left) versus cancer cells (right; Gleason grade 7 or higher) was significant (P < 0.02). HA total of 90 samples were stained with DDX3. B, survival analysis of prostate cancer patients in low and high cytoplasmic DDX3-expressing tumors. (Kaplan–Meier curve and log-rank test, P = 0.630). C, survival analysis of prostate cancer patients in absent and present nuclear DDX3-expressing tumors. (Kaplan–Meier curve and log-rank test, P = 0.359).
Figure 2
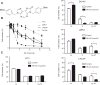
The effect of DDX3 knockdown on prostate cancer colony-forming ability and cell proliferation rate. A, DDX3 expression level of prostate cancer cell lines including PC3, LNCaP, DU145, 22Rv1. B, immunoblot indicating DDX3 knockdown by shDDX3 lentiviral vector. C, normalized colony counts of prostate cancer cell lines DU145, PC3, 22Rv1 transduced with control vector and shDDX3 lentiviral vector, respectively. D, normalized proliferation rate of prostate cancer cell lines DU145, PC3, 22Rv1 transduced with control vector and shDDX3 lentiviral vector.
Figure 3
The effect of RK-33 on cell viability and cell-cycle control of prostate cancer cell lines including PC3, DU145, 22Rv1 and LNCaP. A, the structure of small DDX3 inhibitor RK-33. B, cell viability assay of RK-33 on PC3, DU145, 22Rv1, and LNCaP. C, cell-cycle analysis by flow cytometry of RK-33 on prostate cancer cell lines PC3, DU145, 22Rv1, and LNCaP.
Figure 4
Combination effect of RK-33 treatment and radiation on colony formation of prostate cancer cell lines including PC3, DU145, 22Rv1, and LNCaP.
Figure 5
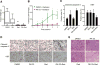
Effect of RK-33 treatment on radiation-induced DNA damage represented by γH2AX foci. A, DU145 cells (20,000 cells per chamber slide) treated with 3 μmol/L RK-33/2 Gy radiation; γH2AX foci per cell were counted for treatment as indicated in the graph. B, LNCaP cells (20,000 cells per chamber slide) treated with 6 μmol/L RK-33/2 Gy radiation. C, PC3 cells (20,000 cells per chamber slide) treated with 12 μmol/L RK-33/2 Gy radiation. D, representative γH2AX foci pictures for three prostate cancer cell lines (DU145, PC3, and LNCaP) with radiation or combination treatment of radiation and RK-33 at time 0, 1, 6, and 24 hours.
Figure 6
Combination treatment in DU145-Luc–inoculated SCID mice reduced tumor growth. A, box plot of tumor weight of four mice groups treated with DMSO, RK-33, radiation, and combination of RK-33 and radiation. B, tumor growth rate of radiation normalized to week 0 indicated by fold change of total flux of bioluminescence emission from live DU145-Luc–inoculated tumors. C, bar graph indicating percentage of nuclei positive for Ki67 (left) and cleaved caspase-3 (right) per tumor per treatment group. D, example of cleaved caspase-3 expression (top) and Ki67 expression (bottom) of each treatment group at 40 magnification. E, H&E staining of tumor slides of each treatment group. Tumors were analyzed 5 weeks after treatment.
Similar articles
- Targeting DDX3 with a small molecule inhibitor for lung cancer therapy.
Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, Levine A, Irving A, Korz D, Tantravedi S, Heerma van Voss MR, Gabrielson K, Bordt EA, Polster BM, Cope L, van der Groep P, Kondaskar A, Rudek MA, Hosmane RS, van der Wall E, van Diest PJ, Tran PT, Raman V. Bol GM, et al. EMBO Mol Med. 2015 May;7(5):648-69. doi: 10.15252/emmm.201404368. EMBO Mol Med. 2015. PMID: 25820276 Free PMC article. - Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer.
Heerma van Voss MR, Vesuna F, Trumpi K, Brilliant J, Berlinicke C, de Leng W, Kranenburg O, Offerhaus GJ, Bürger H, van der Wall E, van Diest PJ, Raman V. Heerma van Voss MR, et al. Oncotarget. 2015 Sep 29;6(29):28312-26. doi: 10.18632/oncotarget.4873. Oncotarget. 2015. PMID: 26311743 Free PMC article. - Targeting mitochondrial translation by inhibiting DDX3: a novel radiosensitization strategy for cancer treatment.
Heerma van Voss MR, Vesuna F, Bol GM, Afzal J, Tantravedi S, Bergman Y, Kammers K, Lehar M, Malek R, Ballew M, Ter Hoeve N, Abou D, Thorek D, Berlinicke C, Yazdankhah M, Sinha D, Le A, Abrahams R, Tran PT, van Diest PJ, Raman V. Heerma van Voss MR, et al. Oncogene. 2018 Jan 4;37(1):63-74. doi: 10.1038/onc.2017.308. Epub 2017 Sep 4. Oncogene. 2018. PMID: 28869602 Free PMC article. - DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs.
Kukhanova MK, Karpenko IL, Ivanov AV. Kukhanova MK, et al. Molecules. 2020 Feb 24;25(4):1015. doi: 10.3390/molecules25041015. Molecules. 2020. PMID: 32102413 Free PMC article. Review. - Targeting host DEAD-box RNA helicase DDX3X for treating viral infections.
Winnard PT Jr, Vesuna F, Raman V. Winnard PT Jr, et al. Antiviral Res. 2021 Jan;185:104994. doi: 10.1016/j.antiviral.2020.104994. Epub 2020 Dec 7. Antiviral Res. 2021. PMID: 33301755 Free PMC article. Review.
Cited by
- DDX3X interacts with SIRT7 to promote PD-L1 expression to facilitate PDAC progression.
Zhao T, Zhu H, Zou T, Zhao S, Zhou L, Ni M, Liu F, Zhu H, Dou X, Di J, Xu B, Wang L, Zou X. Zhao T, et al. Oncogenesis. 2024 Feb 5;13(1):8. doi: 10.1038/s41389-024-00509-2. Oncogenesis. 2024. PMID: 38316768 Free PMC article. - Targeting DDX3X eliminates leukemia stem cells in chronic myeloid leukemia by blocking NT5DC2 mRNA translation.
Duan C, Lin X, Zou W, He Q, Wei F, Pan J, Liu C, Jin Y. Duan C, et al. Oncogene. 2024 Nov 8. doi: 10.1038/s41388-024-03215-w. Online ahead of print. Oncogene. 2024. PMID: 39516658 - DDX3X and Stress Granules: Emerging Players in Cancer and Drug Resistance.
Zhang H, Mañán-Mejías PM, Miles HN, Putnam AA, MacGillivray LR, Ricke WA. Zhang H, et al. Cancers (Basel). 2024 Mar 12;16(6):1131. doi: 10.3390/cancers16061131. Cancers (Basel). 2024. PMID: 38539466 Free PMC article. Review. - Prokaryotic Expression and Affinity Purification of DDX3 Protein.
Huang L, Liang Y, Hou H, Tang M, Liu X, Ma YN, Liang S. Huang L, et al. Protein Pept Lett. 2024;31(3):236-246. doi: 10.2174/0109298665285625231222075700. Protein Pept Lett. 2024. PMID: 38303525 - DEAD-Box RNA Helicases DDX3X and DDX5 as Oncogenes or Oncosuppressors: A Network Perspective.
Secchi M, Lodola C, Garbelli A, Bione S, Maga G. Secchi M, et al. Cancers (Basel). 2022 Aug 6;14(15):3820. doi: 10.3390/cancers14153820. Cancers (Basel). 2022. PMID: 35954483 Free PMC article. Review.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. - PubMed
- Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–34. - PubMed
- Esfahani M, Ataei N, Panjehpour M. Biomarkers for evaluation of prostate cancer prognosis. Asian Pac J Cancer Prev. 2015;16:2601–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical