Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously - PubMed (original) (raw)
Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously
Hongbaek Cho et al. Nat Microbiol. 2016.
Abstract
Multi-protein complexes organized by cytoskeletal proteins are essential for cell wall biogenesis in most bacteria. Current models of the wall assembly mechanism assume that class A penicillin-binding proteins (aPBPs), the targets of penicillin-like drugs, function as the primary cell wall polymerases within these machineries. Here, we use an in vivo cell wall polymerase assay in Escherichia coli combined with measurements of the localization dynamics of synthesis proteins to investigate this hypothesis. We find that aPBP activity is not necessary for glycan polymerization by the cell elongation machinery, as is commonly believed. Instead, our results indicate that cell wall synthesis is mediated by two distinct polymerase systems, shape, elongation, division, sporulation (SEDS)-family proteins working within the cytoskeletal machines and aPBP enzymes functioning outside these complexes. These findings thus necessitate a fundamental change in our conception of the cell wall assembly process in bacteria.
Figures
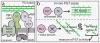
Fig. 1. The Rod system and an in vivo assay of peptidoglycan (PG) polymerase activity
A. Diagram of the currently accepted model for PG biogenesis by the Rod system. Polymers of the actin-like MreB protein organize a complex of membrane proteins including RodA, PBP2, and an aPBP. Glycan polymerization and crosslinking by this complex is thought to be promoted primarily by the peptidoglycan glycosyltransferase (PGT) and transpeptidase (TP) activities of aPBPs with additional TP activity provided by PBP2. B. In untreated cells, PG polymerization and crosslinking by PGT and TP enzymes, respectively are tightly coupled to form the PG matrix (upper panel). When TP activity is inhibited by a beta-lactam, the polymerase working with the blocked TP continues to produce uncrosslinked glycans that are rapidly degraded into fragments that can be isolated and quantified as a measure of polymerase activity (lower panel).
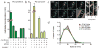
Fig. 2. PG polymerization by the Rod complex does not require aPBP activity
A–B. Cells of HC533(attλC739) [Δ_lysA_ Δ_ampD_ Δ_ponA_ Δ_pbpC_ Δ_mtgA MSponB_ (P_tac_::sulA)] producing SulA to block cell division were pulse labeled with [3H]-mDAP following treatment with the indicated compound(s). Turnover products were extracted with hot water and quantified by HPLC and in-line radiodetection. PG incorporation was determined by digesting the pellets resulting from the hot water extraction with lysozyme and quantifying the amount of label released into the supernatant by scintillation counting. Compound concentrations used were: mecillinam (10 μg/ml), A22 (10 μg/ml), MTSES (1 mM). Results are the average of three independent experiments with the error bars representing the standard error of the mean (SEM). C. Left: Montage with overlaid tracks highlighting MreB movement in HC546(attλHC897) [Δ_ponA_ Δ_pbpC_ Δ_mtgA MSponB (Plac::mreB-SWmNeon)_] after 30 min MTSES inactivation of PBP1b showing continuing MreB motion. Frames 2 s apart, scale bar = 1 μm. Original time-lapse movies are 1 sec/frame. Right top: Kymographs drawn along trajectories indicated on phase contrast image (1, 2, 3, left to right). Each tracked particle is highlighted with a colored trajectory with the color of the track (blue to red) indicating the passage of time. D. Distribution of velocities of MreB motion taken at different points after aPBP inhibition with MTSES (1 mM). For the tracks that we can accurately calculate a particle’s velocity, the fraction of moving particles only declines slightly (from 76% to 66%) during the time course following MTSES treatment. Microscopy results are representative of at least two independent experiments.
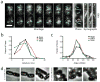
Fig. 3. PBP2 and RodA display directed, circumferential motions similar to MreB
A. Left to right: Montage of PBP2 movement with overlaid tracks in HC596(attHKHC943) [Δ_ponA_ Δ_pbpC_ Δ_mtgA_ Δ_pbpA_ (Plac::msfgfp-pbpA)]. Frames 2 s apart. Each tracked particle is highlighted with a colored trajectory as in Figure 2C. Trajectories 1, 2, and 3 in kymographs are in order left to right. B. Distribution of velocities of tracked particles of MreB (n = 807), PBP2 (n=1234) and RodA (n=243). C. Distribution of angles of PBP2 and RodA trajectories relative to the cell midline. D. Tracked particles of MreB-SWmNeon at 0–30 or 210–240 min after induction of RodA(D262N) from strain TB28(attHKHC929)/pHC938 [WT(PtetA::mreB_-SW_mNeon)/Plac::_pbpA-rodA(D262N)_]. Each tracked particle is highlighted with a different color trajectory overlaid on a phase contrast image. All scale bars are 1 μm. In all cases, original time-lapse movies are 1 sec/frame. Microscopy results are representative of at least two independent experiments.
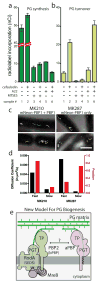
Fig. 4. aPBPs can function independently from the cytoskeletal machinery
A–B. PG matrix assembly and turnover were measured as in Figure 2 using strain HC533(attλC739) [Δ_lysA_ Δ_ampD_ Δ_ponA_ Δ_pbpC_ Δ_mtgA MSponB_ (P_tac_::sulA)]. Cefsulodin was used at 100 μg/ml. Results are the average of three independent experiments with the error bars representing the SEM. C. Tracks of mNeon-PBP1 expressed as (right) the only copy or (left) in addition to native untagged protein in B. subtilis. Each continuously tracked particle is highlighted with a different color trajectory. Note that although no MreB-like directional motion was observed, particles occasionally travel rapidly in one direction for a few frames as expected for membrane diffusion. D. Graph showing diffusion constants, and fraction of particles tracked in each diffusion state as determined by CDF analysis. Microscopy results are representative of at least two independent experiments. E. Schematic view of a new model for PG biogenesis involving two different classes of PG polymerases working semi-autonomously. SEDS PGTs and partner bPBPs perform PG polymerization and crosslinking in the context of the Rod system and divisome (not shown) while aPBPs function outside of these complexes. Collaboration between the synthases likely occurs but the mechanism remains to be defined.
Similar articles
- Discovery of a Diverse Set of Bacteria That Build Their Cell Walls without the Canonical Peptidoglycan Polymerase aPBP.
Atwal S, Chuenklin S, Bonder EM, Flores J, Gillespie JJ, Driscoll TP, Salje J. Atwal S, et al. mBio. 2021 Aug 31;12(4):e0134221. doi: 10.1128/mBio.01342-21. Epub 2021 Jul 27. mBio. 2021. PMID: 34311584 Free PMC article. - Polar Growth in Corynebacterium glutamicum Has a Flexible Cell Wall Synthase Requirement.
Sher JW, Lim HC, Bernhardt TG. Sher JW, et al. mBio. 2021 Jun 29;12(3):e0068221. doi: 10.1128/mBio.00682-21. Epub 2021 Jun 8. mBio. 2021. PMID: 34098735 Free PMC article. - SEDS proteins are a widespread family of bacterial cell wall polymerases.
Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. Meeske AJ, et al. Nature. 2016 Sep 29;537(7622):634-638. doi: 10.1038/nature19331. Epub 2016 Aug 15. Nature. 2016. PMID: 27525505 Free PMC article. - Cell Cycle Machinery in Bacillus subtilis.
Errington J, Wu LJ. Errington J, et al. Subcell Biochem. 2017;84:67-101. doi: 10.1007/978-3-319-53047-5_3. Subcell Biochem. 2017. PMID: 28500523 Free PMC article. Review. - Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis.
Shockman GD, Daneo-Moore L, Kariyama R, Massidda O. Shockman GD, et al. Microb Drug Resist. 1996 Spring;2(1):95-8. doi: 10.1089/mdr.1996.2.95. Microb Drug Resist. 1996. PMID: 9158729 Review.
Cited by
- A Small Molecule Inhibitor of CTP Synthetase Identified by Differential Activity on a Bacillus subtilis Mutant Deficient in Class A Penicillin-Binding Proteins.
Emami K, Wu LJ, Errington J. Emami K, et al. Front Microbiol. 2020 Aug 26;11:2001. doi: 10.3389/fmicb.2020.02001. eCollection 2020. Front Microbiol. 2020. PMID: 32973723 Free PMC article. - Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects.
Vigouroux A, Cordier B, Aristov A, Alvarez L, Özbaykal G, Chaze T, Oldewurtel ER, Matondo M, Cava F, Bikard D, van Teeffelen S. Vigouroux A, et al. Elife. 2020 Jan 6;9:e51998. doi: 10.7554/eLife.51998. Elife. 2020. PMID: 31904338 Free PMC article. - Cell Shape and Antibiotic Resistance Are Maintained by the Activity of Multiple FtsW and RodA Enzymes in Listeria monocytogenes.
Rismondo J, Halbedel S, Gründling A. Rismondo J, et al. mBio. 2019 Aug 6;10(4):e01448-19. doi: 10.1128/mBio.01448-19. mBio. 2019. PMID: 31387909 Free PMC article. - Penicillin-Binding Protein 1 (PBP1) of Staphylococcus aureus Has Multiple Essential Functions in Cell Division.
Wacnik K, Rao VA, Chen X, Lafage L, Pazos M, Booth S, Vollmer W, Hobbs JK, Lewis RJ, Foster SJ. Wacnik K, et al. mBio. 2022 Aug 30;13(4):e0066922. doi: 10.1128/mbio.00669-22. Epub 2022 Jun 15. mBio. 2022. PMID: 35703435 Free PMC article. - MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis.
Hussain S, Wivagg CN, Szwedziak P, Wong F, Schaefer K, Izoré T, Renner LD, Holmes MJ, Sun Y, Bisson-Filho AW, Walker S, Amir A, Löwe J, Garner EC. Hussain S, et al. Elife. 2018 Feb 22;7:e32471. doi: 10.7554/eLife.32471. Elife. 2018. PMID: 29469806 Free PMC article.
References
- McKenna M. Antibiotic resistance: The last resort. Nature. 2013;499:394–396. - PubMed
- Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. - PubMed
- Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. - PubMed
Grants and funding
- R01 AI083365/AI/NIAID NIH HHS/United States
- T32 GM007598/GM/NIGMS NIH HHS/United States
- DP2 AI117923/AI/NIAID NIH HHS/United States
- R01 AI099144/AI/NIAID NIH HHS/United States
- U19 AI109764/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous