Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization - PubMed (original) (raw)
. 2016 Sep 26;214(7):817-30.
doi: 10.1083/jcb.201601071. Epub 2016 Sep 19.
Mari Mito 2, Satoshi Kurosaka 3, Toru Takumi 3, Chiharu Tanegashima 4, Takeshi Chujo 5, Kaori Yanaka 2, Robert E Kingston 1, Tetsuro Hirose 5, Charles Bond 6, Archa Fox 7, Shinichi Nakagawa 8
Affiliations
- PMID: 27646274
- PMCID: PMC5037409
- DOI: 10.1083/jcb.201601071
Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization
Jason A West et al. J Cell Biol. 2016.
Abstract
Paraspeckles are nuclear bodies built on the long noncoding RNA Neat1, which regulates a variety of physiological processes including cancer progression and corpus luteum formation. To obtain further insight into the molecular basis of the function of paraspeckles, we performed fine structural analyses of these nuclear bodies using structural illumination microscopy. Notably, paraspeckle proteins are found within different layers along the radially arranged bundles of Neat1 transcripts, forming a characteristic core-shell spheroidal structure. In cells lacking the RNA binding protein Fus, paraspeckle spheroids are disassembled into smaller particles containing Neat1, which are diffusely distributed in the nucleoplasm. Sequencing analysis of RNAs purified from paraspeckles revealed that AG-rich transcripts associate with Neat1, which are distributed along the shell of the paraspeckle spheroids. We propose that paraspeckles sequester core components inside the spheroids, whereas the outer surface associates with other components in the nucleoplasm to fulfill their function.
© 2016 West et al.
Figures
Figure 1.
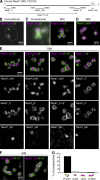
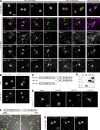
Core-shell arrangement of Neat1 in paraspeckle spheres I. (A) Schematic diagrams of the positions of FISH probes that detect differential regions of Neat1. (B and C) Simultaneous detection of the middle and 3′ regions of Neat1 using a conventional epifluorescence microscope (Conventional) and SIM. (D) The same FISH image detected with the converse combination of secondary antibodies as in C. (E) Comparisons of the differential distribution of each Neat1 region in the paraspeckle spheres. Note that the middle region is located in the core of the paraspeckles, whereas the 5′ and the 3′ regions are located peripherally. Asterisks indicate the position of the putative transcription site detected with the Neat1_tail probe. (F) Paraspeckles with a sausage-like shape that were occasionally detected in the corpus luteal cells. (G) Histogram of paraspeckles with different shapes (n = 187). Bars: (B) 5 µm; (C–F) 500 nm.
Figure 2.
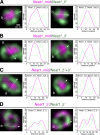
Core-shell arrangement of Neat1 in paraspeckle spheres II. Higher magnification SIM images of two of the representative single paraspeckles stained with the Neat1_mid and the Neat1_5′ probe (A), the Neat1_mid and the Neat1_3′ probe (B), the Neat1_mid and the Neat1_5′+3′ probe (C), and the _Neat1_3_′ and the Neat1_ 5_′ probe (D). Intensity profiles along the dashed lines (a and b) are shown in the graphs. Note that the middle region of Neat1 is centrally located, and the 5_′ and the 3′ regions are distributed in a complementary manner along the shell of the paraspeckle spheres. Bar, 100 nm.
Figure 3.
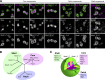
Core-shell arrangement of protein components in paraspeckle spheres I. (A) Simultaneous detection of Neat1 and seven of the protein components of paraspeckles, including Sfpq, Nono, Pspc1, Fus, Rbm14, Brg1, and Tardbp in corpus luteal cells. Note that the paraspeckle proteins are grouped into the core, patch, and shell components depending on their distribution in the paraspeckles. (B) Dendrogram based on pairwise class-distance matrix generated using the machine-learning pattern-recognition tool wndchrm. The shell, core, and patch components are grouped into three distinct branches. (C) A model for the structure of paraspeckles. Neat1 folds in half with the 5′ and the 3′ regions bundled independently and radially arranged to construct scaffolds of paraspeckles. Bar, 500 nm.
Figure 4.
Core-shell arrangement of protein components in paraspeckle spheres II. Higher magnification SIM images of two of the representative single paraspeckles stained with the _Neat1 5′+3_′ probe and Sfpq (A), Nono (B), Pspc1 (C), Fus (D), Rbm14 (E), Brg1 (F), and Tardbp (G). Intensity profiles along the dashed lines (a and b) are shown in the graphs next to the images. Bar, 100 nm.
Figure 5.
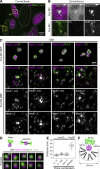
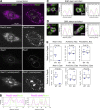
Fus is required for the assembly of the core-shell structure of paraspeckles. (A) Simultaneous detection of Fus and Neat1 in a mixture of MEFs prepared from WT and Fus KO mice using a conventional epifluorescence microscope (Conventional). Yellow boxes indicate the areas shown at a higher magnification in B. Note that Neat1 forms discrete nuclear body-like structures in Fus KO MEFs. (B) Higher magnification images shown in yellow boxes in A. (C) SIM observation of Neat1 in WT and Fus KO MEFs using the region-specific probes. Note that the characteristic core-shell structure was not observed in the Fus KO MEFs. Asterisks indicate the position of the putative transcription sites detected with the Neat1 tail probe. Arrowheads indicate Neat1 primary units containing both of the detected regions of Neat1. (D) Models of the Neat1 primary unit and higher magnification images of FISH signals obtained with the Neat1_5′+3′ probe in Fus KO MEFs. Note the close association of the two signals. (E) Measurement of the distance between the two different regions of Neat1 in Fus KO MEFs. Note that the distance between the 5′ and 3′ region of Neat1 is shorter than the distance between the 5′ and the middle or the middle and the 3′ regions of Neat1. The median is indicated with a horizontal line in a box that represents the first and third quartiles. Outliers are indicated as circles, and the maximum and minimums are indicated at the end of the whiskers. Each dot represents each signal of Neat1 particle. (F) A model of the organization of Neat1 in paraspeckle spheres. Bars: (A) 10 µm; (B) 1 µm; (C) 500 nm; (D) 100 nm.
Figure 6.
Fus-independent and dependent recruitment of paraspeckle proteins. (A) Simultaneous detection of Neat1 and seven of the protein components of paraspeckles, including Sfpq, Nono, Pspc1, Fus, Rbm14, Brg1, and Tardbp, in MEFs derived from WT and Fus KO mice. Note that DBHS family proteins (Sfpq, Nono, and Pspc1) and Tardbp, but not Rbm14 and Brg1, are recruited to the putative transcription site in the absence of Fus. Arrowheads indicate paraspeckle-like nuclear bodies formed at the putative Neat1 transcription site in Fus KO MEFs. (B) Simultaneous detection of various forms of NEAT1 and NONO in HAP1 cells and FUS-deleted HAP1 cells (ΔFUS HAP1). Probes used to detect NEAT1 are shown in the top boxes. (C) Schematic drawing of full-length and mutant FUS protein exogenously expressed by lentiviruses. ΔN FUS lack the PrLD and ΔC FUS lack the RNA binding domains including RNA recognition motifs (RRM) and arginine (R)-glycine-glycine domain (RGG) as well as zinc finger domain (ZF). (D) Western blot analyses of lysate from the cells infected with control EGFP (C), full-length FUS (FL), ΔN FUS (ΔN), and ΔC FUS (ΔC). Note that migration of FL and ΔN are much slower than predicted molecular mass (57 and 35 kD, respectively), probably because of the presence of PrLD in these molecules. (E) Simultaneous detection of Neat1 5′+3′ and Nono in Fus KO MEFs expressing various forms of FUS protein. Note that the core-shell structure of paraspeckles was rescued with FL FUS, but not with mutant molecules that lack either PrLD or RNA binding domains. (F) Confirmation of the specificity of polyclonal [Fus (poly)] and monoclonal (Fus) antibodies against Fus. Mixtures of MEFs derived from WT and KO mice of Fus were stained with each antibody. Note the complete absence of signals in the Fus KO MEFs (arrowheads). The positions of the epitope of these antibodies are shown in the schematic drawing of the domain structure of Fus. (G) Simultaneous detection of Fus using polyclonal antibodies and mAbs that recognize the N- and C-terminal region of the protein, respectively. Bars: (A, B, E, and G) 500 nm; (F) 200 µm.
Figure 7.
Identification of novel paraspeckle RNA components by CHART RNAseq. (A) Schematic cartoon showing the CHART purification of paraspeckle fragments. The Neat1 complexes were purified using antisense oligonucleotides, and copurified RNAs were analyzed by RNAseq. (B) Schematic of the Neat1 locus showing the position of the oligonucleotide sets (oligos_A and oligos_B) used for the CHART purification and mapped reads of the input and CHART-purified RNAs. The scales are automatically adjusted in the top panel and adjusted to a distinct value (0–2,000) in the bottom panel. Note that the 5′ region of Neat1 is predominantly enriched by the CHART purification, whereas the 3′ region is also moderately enriched by both oligonucleotide sets. Mapping of CHART-enriched RNAseq reads at the genomic loci of Trim44, Numa1, Actr3, and Prss35. Note that the reads are mapped to exons in Trim44 and Numa1 (C), whereas they are mapped to the third and the first intron of Actr3 and Prss35, respectively (D). MEME-identified AG-rich sequence motifs and their distribution along the exon-enriched (E) and intron-enriched (F) genes. Partial regions of each intron containing the AG-rich motifs are shown in F. Bar, 500 nm.
Figure 8.
FISH analyses of the localization of paraspeckle-enriched AG-rich RNAs. (A and B) Simultaneous detection of Neat1 and the exons and the first intron of Prss35 in corpus luteal cells using confocal microscopy. A single optical section image is shown. Note that subpopulation of Prss35 intron signals colocalized with Neat1-positive paraspeckles, whereas exon signals were mostly observed in the cytoplasm and did not coincide with the Neat1 signals. Intensity profiles along the yellow dashed line are shown in B. Dashed white curving lines indicate position of the nucleus. Note that the bright round signals in the cytoplasm are derived from autofluorescence of lipid droplets, some of which are shown by asterisks and are clearly identifiable in a different channel overexposed for Neat1 signals. Bar, 10 µm. (C and D) Simultaneous detection of AG-rich RNA and Neat1_5′+3′ in corpus luteal cells using SIM. Intensity profiles along the dashed line are shown in the graphs adjacent to the images. Bars, 500 nm. (E) Box and whisker plots showing the expression of Neat1 and AG-rich RNAs in the cytoplasm (cyto) or nucleus (nuc) of corpus luteal cells from WT and Neat1 KO mice. The median is indicated with a horizontal line in a box that represents the first and the third quartiles. Outliers are indicated as circles, and the maximum and the minimums are indicated at the end of the whiskers. Each blue dot represents a sample from an individual mouse.
Similar articles
- Organization and function of paraspeckles.
Wang Y, Chen LL. Wang Y, et al. Essays Biochem. 2020 Dec 7;64(6):875-882. doi: 10.1042/EBC20200010. Essays Biochem. 2020. PMID: 32830222 Review. - ALS-linked FUS mutations confer loss and gain of function in the nucleus by promoting excessive formation of dysfunctional paraspeckles.
An H, Skelt L, Notaro A, Highley JR, Fox AH, La Bella V, Buchman VL, Shelkovnikova TA. An H, et al. Acta Neuropathol Commun. 2019 Jan 14;7(1):7. doi: 10.1186/s40478-019-0658-x. Acta Neuropathol Commun. 2019. PMID: 30642400 Free PMC article. - Shedding light on paraspeckle structure by super-resolution microscopy.
Hu SB, Yao RW, Chen LL. Hu SB, et al. J Cell Biol. 2016 Sep 26;214(7):789-91. doi: 10.1083/jcb.201609008. Epub 2016 Sep 19. J Cell Biol. 2016. PMID: 27646270 Free PMC article. - Compromised paraspeckle formation as a pathogenic factor in FUSopathies.
Shelkovnikova TA, Robinson HK, Troakes C, Ninkina N, Buchman VL. Shelkovnikova TA, et al. Hum Mol Genet. 2014 May 1;23(9):2298-312. doi: 10.1093/hmg/ddt622. Epub 2013 Dec 11. Hum Mol Genet. 2014. PMID: 24334610 Free PMC article. - Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles.
Hirose T, Yamazaki T, Nakagawa S. Hirose T, et al. Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1545. doi: 10.1002/wrna.1545. Epub 2019 May 1. Wiley Interdiscip Rev RNA. 2019. PMID: 31044562 Review.
Cited by
- ArcRNAs and the formation of nuclear bodies.
Nakagawa S, Yamazaki T, Mannen T, Hirose T. Nakagawa S, et al. Mamm Genome. 2022 Jun;33(2):382-401. doi: 10.1007/s00335-021-09881-5. Epub 2021 Jun 3. Mamm Genome. 2022. PMID: 34085114 Review. - TDP-43 Proteinopathy and Tauopathy: Do They Have Pathomechanistic Links?
Riku Y, Yoshida M, Iwasaki Y, Sobue G, Katsuno M, Ishigaki S. Riku Y, et al. Int J Mol Sci. 2022 Dec 12;23(24):15755. doi: 10.3390/ijms232415755. Int J Mol Sci. 2022. PMID: 36555399 Free PMC article. Review. - Involvement of the long noncoding RNA NEAT1 in carcinogenesis.
Klec C, Prinz F, Pichler M. Klec C, et al. Mol Oncol. 2019 Jan;13(1):46-60. doi: 10.1002/1878-0261.12404. Epub 2018 Dec 3. Mol Oncol. 2019. PMID: 30430751 Free PMC article. Review. - Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu.
Nakagawa S, Yamazaki T, Hirose T. Nakagawa S, et al. Open Biol. 2018 Oct 24;8(10):180150. doi: 10.1098/rsob.180150. Open Biol. 2018. PMID: 30355755 Free PMC article. Review. - Self-assembly of promoter DNA and RNA Pol II machinery into transcriptionally active biomolecular condensates.
Lewis BA, Das SK, Jha RK, Levens D. Lewis BA, et al. Sci Adv. 2023 Oct 20;9(42):eadi4565. doi: 10.1126/sciadv.adi4565. Epub 2023 Oct 18. Sci Adv. 2023. PMID: 37851801 Free PMC article.
References
- Cerase A., Smeets D., Tang Y.A., Gdula M., Kraus F., Spivakov M., Moindrot B., Leleu M., Tattermusch A., Demmerle J., et al. . 2014. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc. Natl. Acad. Sci. USA. 111:2235–2240. 10.1073/pnas.1312951111 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources