Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes - PubMed (original) (raw)
. 2016 Oct 11;24(4):593-607.
doi: 10.1016/j.cmet.2016.08.020. Epub 2016 Sep 22.
Athanasia Palasantza 2, Pernilla Eliasson 3, Eva-Marie Andersson 3, Anne-Christine Andréasson 3, Xiaoyan Sun 4, Simone Picelli 5, Alan Sabirsh 3, Maryam Clausen 6, Magnus K Bjursell 7, David M Smith 8, Maria Kasper 4, Carina Ämmälä 3, Rickard Sandberg 9
Affiliations
- PMID: 27667667
- PMCID: PMC5069352
- DOI: 10.1016/j.cmet.2016.08.020
Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes
Åsa Segerstolpe et al. Cell Metab. 2016.
Abstract
Hormone-secreting cells within pancreatic islets of Langerhans play important roles in metabolic homeostasis and disease. However, their transcriptional characterization is still incomplete. Here, we sequenced the transcriptomes of thousands of human islet cells from healthy and type 2 diabetic donors. We could define specific genetic programs for each individual endocrine and exocrine cell type, even for rare δ, γ, ε, and stellate cells, and revealed subpopulations of α, β, and acinar cells. Intriguingly, δ cells expressed several important receptors, indicating an unrecognized importance of these cells in integrating paracrine and systemic metabolic signals. Genes previously associated with obesity or diabetes were found to correlate with BMI. Finally, comparing healthy and T2D transcriptomes in a cell-type resolved manner uncovered candidates for future functional studies. Altogether, our analyses demonstrate the utility of the generated single-cell gene expression resource.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
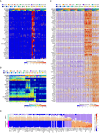
Single-Cell Transcriptome Analyses of Human Pancreas (A) Table of donor information (HbA1c, glycated hemoglobin). (B) Projection of all cells (n = 2,209) onto two dimensions using t-SNE based on the expression values (log2RPKM) of the 1,000 genes with highest biological variation across cells. (C) Expression (log2RPKM) of exocrine marker genes (PRSS1 for acinar and KRT19 for ductal cells) overlaid onto the 2D t-SNE as shown in (B). (D) Boxplots displaying the expression levels in the seven obtained clusters of marker genes for MHC class II antigen-presenting cells (CD86), mast cells (TPSAB1), pancreatic stellate cells (COL1A2), and endothelial cells (PLVAP). Median and mean are shown as a line and circle, respectively. Edges of each box indicate the 25th and 75th percentiles. Bars extend to extreme data points and outliers are plotted as gray dots. (E) Two-dimensional t-SNE projection of the endocrine cells (n = 1,554) based on the expression values (log2RPKM) of the 500 genes with highest biological variation across endocrine cells. The obtained clusters were assigned to the endocrine cell types based on the hormone expression levels in (F). Colors correspond to cell types and shadings indicate donors. Healthy and T2D cells are marked with circles and triangles, respectively. (F) t-SNE representation of cells as in (E) illustrating the expression of the five endocrine hormones: GCG, INS, PPY, SST, and GHRL. Color scale is according to log2RPKM values, with white and red colors corresponding to minimum (zero) and maximum (log2RPKM = 21) expression, respectively. (G) Bar graphs showing the percentage of cells classified into cell types, per donor.
Figure 2
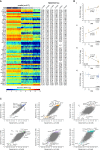
Characterization of Endocrine Cell Transcriptomes (A) Boxplots showing the number of genes detected in each cell type (expression threshold, RPKM ≥ 1). Median and mean are shown as a line and circle, respectively. Edges of each box indicate the 25th and 75th percentiles. Bars extend to extreme data points and outliers are plotted as gray dots. (B) Percentage of all mRNAs in respect to the total transcriptome in each cell type (Table S1), with genes ranked according to the expression magnitude in descending order (x axis). (C) Table with the number of significantly enriched genes in α, β, γ, δ, acinar, and ductal cells. (D) Heatmap with expression distributions for the top 25 enriched genes in each of the four endocrine cell types (α, β, γ, and δ cells). The genes were selected based on the magnitude of expression range among the four endocrine cell types. The expression profiles of the ten donors are shown separately and for each endocrine and exocrine cell type, with labels indicating cell type and donor (top). Colors correspond to standardized log2 expression values, where each cell in the heatmap contains the distribution of values across the cells for each cell type and donor. (E) Boxplots with the expression levels of selected cell-type enriched genes identified in the differential expression analysis: FAP, DPP4, and GPR119 for α cells; LINC01099 for β cells; and LEPR and GHSR for δ cells. Gene expression is shown for α, β, γ, δ, ε, acinar, and ductal cells, with color shadings representing different donors. Median and mean are shown as a line and circle, respectively. Edges of each box indicate the 25th and 75th percentiles. Bars extend to extreme data points and outliers are plotted as gray dots. (F and G) Single-molecule RNA FISH on pancreatic tissue section. (F, left) A representative islet (donor H5) co-stained with GCG (green), FAP (red), and DAPI (blue). (G, left) Islet (donor H5) co-stained with SST (green), LEPR (red), and DAPI (blue). (F and G, right) Zoom-in on merged or individual channels. Quantification and additional images in Table S3. Scale bar represents 25 μm.
Figure 3
Cell-Type-Specific Expression (A) Heatmap showing ε cell enriched gene expression (rows) across the seven cell types for the ten donors (columns). Labels indicating the cell type and donor are shown on the top. Colors in the heatmap correspond to standardized log2 expression values, where each cell in the heatmap contains the distribution of values across the cells for each cell type and donor. (B) Heatmap with expression levels of transcription factors (rows) across the cells from the six cell types for the ten donors (columns). Blue and red colors correspond to minimum (zero) and maximum (log2RPKM = 12) expression, respectively. (C) Heatmap with expression of significantly enriched genes (rows) in acinar and ductal cells across the six cell types for the ten donors (columns). The genes were selected based on the magnitude of expression range between the two exocrine cell types. The heatmap was generated as in (A). (D) Heatmap showing the differentially expressed (DE) genes (columns) between the pancreatic stellate (PSCs) and the endothelial cells (rows). The results were obtained using single-cell differential expression (SCDE), and only the genes with a log2 fold change in expression of at least seven between the two groups are shown. The colors in the heatmap correspond to standardized log2 expression values, where each cell in the heatmap contains the distribution of values across the cells of each cell type.
Figure 4
Uncovering Subtypes of Endocrine and Exocrine Cells (A) (Left) Two-dimensional t-SNE representation of all α cells (n = 886, 10 donors) using donor-normalized expression values of the 500 most variable genes in α cells. Colored according to cluster assignments. (Right) Heatmap illustrating the top DE genes (columns) between the two α cell clusters (rows). The colors in the heatmap correspond to standardized log2 expression values, where each cell in heatmap contains the distribution of values across the cells in each cluster. (B) (Left) Two-dimensional t-SNE representation of β cells (n = 270, 10 donors) using donor-normalized expression values of the 50 most variable genes in β cells. (Right) t-SNE representation of cells, colored according to expression (log2RPKM) of DE genes among the five clusters. (C) (Left) Two-dimensional t-SNE projection of acinar cells (n = 185, 10 donors) using donor-normalized expression values of the 100 most variable genes in acinar cells. (Right) Heatmaps illustrating the top DE genes (columns) per cell cluster (rows). The heatmap was generated as in (A).
Figure 5
Gene Expression Correlates to Donor Physiological Characteristics (A) Heatmap of gene expression associated with BMI in α cells (n = 417, healthy male donors). Genes (rows) with a positive or negative correlation of at least 0.7 in magnitude are ranked in descending order of coefficient. Cells (columns) are ordered in descending order of BMI. Blue and red colors correspond to minimum (zero) and maximum (log2RPKM = 14) expression, respectively. The corresponding Spearman’s correlation coefficients computed for the genes in each cell type (based on the cells from healthy male donors) are displayed on the right of the heatmap. (B) Scatterplots showing the expression levels of four genes with a robust correlation with BMI. The fitted curve in each scatter shows the linear regression between the expression and BMI, where m is the slope and b the y-intercept. (C) Scatterplots of Spearman’s correlations of genes toward BMI computed based on overall expression in donors (x axis) or for the specific indicated cell type (y axis). Colored are the genes with an absolute correlation coefficient of at least 0.6 within a particular cell type that is also at least 10% higher in respect to the correlation computed using all cells.
Figure 6
Altered Gene Expression in Cells from T2D Individuals (A) Bar graphs showing the number of DE genes between cells from healthy and T2D donors per cell type. (B) Heatmaps with the DE genes between healthy and T2D α (left) and β cells (right). Labels indicating the disease status and sex of the cells are shown on the top. Colors in the heatmap correspond to standardized log2 expression values, where each cell in the heatmap contains the distribution of values across the cells of each group. (C) Heatmap of the enriched gene sets (using GSEA) within the α, β, γ, δ, acinar, and ductal cell types for healthy and T2D groups. The heatmap is colored according to the adjusted p values (−log10), with red and blue colors corresponding to enrichments among genes up- and downregulated in T2D, respectively. Sets with no significant enrichment are indicated with white color.
Figure 7
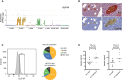
Functional Analysis of GLP1R (A) Boxplot showing expression of GLP1R in α, β, γ, δ, ε, acinar, and ductal cells, with color shadings representing different donors. Median and mean are shown as a line and circle, respectively. Edges of each box indicate the 25th and 75th percentiles. Bars extend to extreme data points and outliers are plotted as gray dots. (B) Immunohistochemistry of pancreatic tissue for INS (brown) and GCG (red) (top left and zoom-in on top right), or GLP1R (brown) (lower left and zoom-in on lower right). (C) (Left) FACS analysis of islet cells stained with fluorescently conjugated GLP1R antagonist. Gate shows fraction of the total islet cells positive for GLP1R. (Right) Pie charts with the percentage of α, β, δ, and other cells in FACS analyses of total islet cells (upper) or the GLP1R+ cellular fraction (lower), labeled with INS, GCG, and SST antibodies. (D) Dot plots showing GSIS of human islets from one healthy (left) and one T2D (right) donor treated with 16.7 mM glucose ± 10 nM Exenatide (Ex4).
Comment in
- Single-Cell Sequencing of Human Pancreatic Islets-New Kids on the Block.
Prasad RB, Groop L. Prasad RB, et al. Cell Metab. 2016 Oct 11;24(4):523-524. doi: 10.1016/j.cmet.2016.09.012. Cell Metab. 2016. PMID: 27732832 - Diabetes: Transcriptomes reveal specific islet cell signatures.
Geach T. Geach T. Nat Rev Endocrinol. 2016 Dec;12(12):686. doi: 10.1038/nrendo.2016.173. Epub 2016 Oct 14. Nat Rev Endocrinol. 2016. PMID: 27739516 No abstract available.
Similar articles
- Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes.
Lawlor N, George J, Bolisetty M, Kursawe R, Sun L, Sivakamasundari V, Kycia I, Robson P, Stitzel ML. Lawlor N, et al. Genome Res. 2017 Feb;27(2):208-222. doi: 10.1101/gr.212720.116. Epub 2016 Nov 18. Genome Res. 2017. PMID: 27864352 Free PMC article. - Single-cell ATAC-Seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures.
Rai V, Quang DX, Erdos MR, Cusanovich DA, Daza RM, Narisu N, Zou LS, Didion JP, Guan Y, Shendure J, Parker SCJ, Collins FS. Rai V, et al. Mol Metab. 2020 Feb;32:109-121. doi: 10.1016/j.molmet.2019.12.006. Epub 2019 Dec 20. Mol Metab. 2020. PMID: 32029221 Free PMC article. - Single-cell transcriptomics of human islet ontogeny defines the molecular basis of β-cell dedifferentiation in T2D.
Avrahami D, Wang YJ, Schug J, Feleke E, Gao L, Liu C; HPAP Consortium; Naji A, Glaser B, Kaestner KH. Avrahami D, et al. Mol Metab. 2020 Dec;42:101057. doi: 10.1016/j.molmet.2020.101057. Epub 2020 Jul 30. Mol Metab. 2020. PMID: 32739450 Free PMC article. - Interrogating islets in health and disease with single-cell technologies.
Carrano AC, Mulas F, Zeng C, Sander M. Carrano AC, et al. Mol Metab. 2017 May 5;6(9):991-1001. doi: 10.1016/j.molmet.2017.04.012. eCollection 2017 Sep. Mol Metab. 2017. PMID: 28951823 Free PMC article. Review. - Genomics of Islet (Dys)function and Type 2 Diabetes.
Lawlor N, Khetan S, Ucar D, Stitzel ML. Lawlor N, et al. Trends Genet. 2017 Apr;33(4):244-255. doi: 10.1016/j.tig.2017.01.010. Epub 2017 Feb 27. Trends Genet. 2017. PMID: 28245910 Free PMC article. Review.
Cited by
- A Comparison for Dimensionality Reduction Methods of Single-Cell RNA-seq Data.
Xiang R, Wang W, Yang L, Wang S, Xu C, Chen X. Xiang R, et al. Front Genet. 2021 Mar 23;12:646936. doi: 10.3389/fgene.2021.646936. eCollection 2021. Front Genet. 2021. PMID: 33833778 Free PMC article. - Altered microvasculature in pancreatic islets from subjects with type 1 diabetes.
Granlund L, Hedin A, Korsgren O, Skog O, Lundberg M. Granlund L, et al. PLoS One. 2022 Oct 31;17(10):e0276942. doi: 10.1371/journal.pone.0276942. eCollection 2022. PLoS One. 2022. PMID: 36315525 Free PMC article. - Evidence for the existence and potential roles of intra-islet glucagon-like peptide-1.
Campbell SA, Johnson J, Light PE. Campbell SA, et al. Islets. 2021 Mar 4;13(1-2):32-50. doi: 10.1080/19382014.2021.1889941. Epub 2021 Mar 16. Islets. 2021. PMID: 33724156 Free PMC article. Review. - Diabetes: Transcriptomes reveal specific islet cell signatures.
Geach T. Geach T. Nat Rev Endocrinol. 2016 Dec;12(12):686. doi: 10.1038/nrendo.2016.173. Epub 2016 Oct 14. Nat Rev Endocrinol. 2016. PMID: 27739516 No abstract available. - Anti-correlated feature selection prevents false discovery of subpopulations in scRNAseq.
Tyler SR, Lozano-Ojalvo D, Guccione E, Schadt EE. Tyler SR, et al. Nat Commun. 2024 Jan 24;15(1):699. doi: 10.1038/s41467-023-43406-9. Nat Commun. 2024. PMID: 38267438 Free PMC article.
References
- Braks J.A., Martens G.J. 7B2 is a neuroendocrine chaperone that transiently interacts with prohormone convertase PC2 in the secretory pathway. Cell. 1994;78:263–273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical