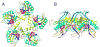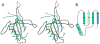Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems - PubMed (original) (raw)
Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems
Andrei Fokine et al. Structure. 2016.
Abstract
Many large viruses, including tailed dsDNA bacteriophages and herpesviruses, assemble their capsids via formation of precursors, called procapsids or proheads. The prohead has an internal core, made of scaffolding proteins, and an outer shell, formed by the major capsid protein. The prohead usually contains a protease, which is activated during capsid maturation to destroy the inner core and liberate space for the genome. Here, we report a 2.0 Å resolution structure of the pentameric procapsid protease of bacteriophage T4, gene product (gp)21. The structure corresponds to the enzyme's pre-active state in which its N-terminal region blocks the catalytic center, demonstrating that the activation mechanism involves self-cleavage of nine N-terminal residues. We describe similarities and differences between T4 gp21 and related herpesvirus proteases. We found that gp21 and the herpesvirus proteases have similarity with proteins forming the tubes of phage tails and bacterial type VI secretion systems, suggesting their common evolutionary origin.
Keywords: bacterial type VI secretion system; bacteriophage; herpesvirus; phage tail tube; procapsid; prohead protease; self-cleavage; virus assembly.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
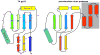
Figure 1. Crystal structure of the T4 prohead protease pentamer
The polypeptide chains, shown as ribbons, are rainbow-colored from blue at the N-terminus to red at the C-terminus. Positions of the catalytic centers are shown by semitransparent spheres. (A) View along the five-fold axis of the pentamer. (B) View perpendicular to the five-fold axis.
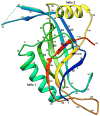
Figure 2. Ribbon diagram of the gp21 monomer rainbow-colored from blue at the N-terminus to red at the C-terminus
Side chains of the residues 140 (Ser of the catalytic triad mutated to Ala), 85 (His of the triad mutated to Ala), and 168 (Asp of the catalytic triad) are shown in black.
Figure 3. Stereo view of the gp21 substrate binding site blocked by the N-terminal region of the polypeptide chain
The peptide bond between Glu9 and Thr10 is located near the side chain of residue 140, which is the catalytic Ser mutated to Ala.
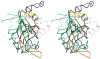
Figure 4. Stereo view of superposition of T4 gp21 with the pseudorabies virus protease
The polypeptide chain of gp21 is rainbow-colored from blue at the N-terminus to red at the C- terminus. The polypeptide chain of the pseudorabies virus protease is shown in black. The C- terminal helical region of the pseudorabies virus protease, absent in gp21, is semitransparent. Residue numbers for the gp21 protein are shown in magenta.
Figure 5. Schematic diagrams showing topology of T4 gp21 (left) and the pseudorabies herpesvirus protease (right)
β–strands are represented by arrows, and α-helices are represented by cylinders. The C-terminal helical region of the pseudorabies virus protease, absent in gp21, is outlined by a grey rectangle.
Figure 6. Comparison of the T4 capsid assembly protease, gp21, with the T4 tail tube protein gp19
(A) Stereo view of the superposition of gp21 with gp19 (PDB ID: 5iv5). Region 10 - 116 of gp21 is rainbow–colored from blue at the N-terminus to yellow-green at the C- terminus. Region 36 – 160 of gp19 is shown in black. Residue numbers for the gp21 chain are shown in magenta. (B) Schematic diagram showing topology of the conserved structural motif. See also Figure S1
Similar articles
- Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease.
Cheng H, Shen N, Pei J, Grishin NV. Cheng H, et al. Protein Sci. 2004 Aug;13(8):2260-9. doi: 10.1110/ps.04726004. Protein Sci. 2004. PMID: 15273316 Free PMC article. - Capsids and Portals Influence Each Other's Conformation During Assembly and Maturation.
Maurer JB, Oh B, Moyer CL, Duda RL. Maurer JB, et al. J Mol Biol. 2020 Mar 27;432(7):2015-2029. doi: 10.1016/j.jmb.2020.01.022. Epub 2020 Feb 6. J Mol Biol. 2020. PMID: 32035900 Free PMC article. - A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly.
D'Lima NG, Teschke CM. D'Lima NG, et al. J Virol. 2015 Oct;89(20):10569-79. doi: 10.1128/JVI.01629-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269173 Free PMC article. - The amazing HK97 fold: versatile results of modest differences.
Duda RL, Teschke CM. Duda RL, et al. Curr Opin Virol. 2019 Jun;36:9-16. doi: 10.1016/j.coviro.2019.02.001. Epub 2019 Mar 8. Curr Opin Virol. 2019. PMID: 30856581 Free PMC article. Review. - Assemblins as maturational proteases in herpesviruses.
Zühlsdorf M, Hinrichs W. Zühlsdorf M, et al. J Gen Virol. 2017 Aug;98(8):1969-1984. doi: 10.1099/jgv.0.000872. Epub 2017 Jul 31. J Gen Virol. 2017. PMID: 28758622 Review.
Cited by
- Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Xiao H, Zhou J, Yang F, Liu Z, Song J, Chen W, Liu H, Cheng L. Xiao H, et al. Viruses. 2023 Jan 26;15(2):355. doi: 10.3390/v15020355. Viruses. 2023. PMID: 36851569 Free PMC article. - Pseudomonas Phage MD8: Genetic Mosaicism and Challenges of Taxonomic Classification of Lambdoid Bacteriophages.
Evseev P, Lukianova A, Sykilinda N, Gorshkova A, Bondar A, Shneider M, Kabilov M, Drucker V, Miroshnikov K. Evseev P, et al. Int J Mol Sci. 2021 Sep 26;22(19):10350. doi: 10.3390/ijms221910350. Int J Mol Sci. 2021. PMID: 34638693 Free PMC article. - Halobacterium salinarum virus ChaoS9, a Novel Halovirus Related to PhiH1 and PhiCh1.
Dyall-Smith M, Palm P, Wanner G, Witte A, Oesterhelt D, Pfeiffer F. Dyall-Smith M, et al. Genes (Basel). 2019 Mar 1;10(3):194. doi: 10.3390/genes10030194. Genes (Basel). 2019. PMID: 30832293 Free PMC article. - Origin, Evolution and Diversity of φ29-like Phages-Review and Bioinformatic Analysis.
Evseev P, Gutnik D, Evpak A, Kasimova A, Miroshnikov K. Evseev P, et al. Int J Mol Sci. 2024 Oct 9;25(19):10838. doi: 10.3390/ijms251910838. Int J Mol Sci. 2024. PMID: 39409167 Free PMC article. Review. - Protein interactions and consensus clustering analysis uncover insights into herpesvirus virion structure and function relationships.
Hernández Durán A, Greco TM, Vollmer B, Cristea IM, Grünewald K, Topf M. Hernández Durán A, et al. PLoS Biol. 2019 Jun 14;17(6):e3000316. doi: 10.1371/journal.pbio.3000316. eCollection 2019 Jun. PLoS Biol. 2019. PMID: 31199794 Free PMC article.
References
- Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–243. - PubMed
- Akhter T, Zhao L, Kohda A, Mio K, Kanamaru S, Arisaka F. The neck of bacteriophage T4 is a ring-like structure formed by a hetero-oligomer of gp13 and gp14. Biochim Biophys Acta. 2007;1774:1036–1043. - PubMed
- Anderson D, Reilly B. Morphogenesis of bacteriophage phi29. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Washington, DC: American Society for Microbiology; 1993. pp. 859–867.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources