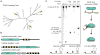Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection - PubMed (original) (raw)
. 2016 Oct 13;538(7624):270-273.
doi: 10.1038/nature19802. Epub 2016 Sep 26.
Affiliations
- PMID: 27669025
- PMCID: PMC5576363
- DOI: 10.1038/nature19802
Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection
Alexandra East-Seletsky et al. Nature. 2016.
Abstract
Bacterial adaptive immune systems use CRISPRs (clustered regularly interspaced short palindromic repeats) and CRISPR-associated (Cas) proteins for RNA-guided nucleic acid cleavage. Although most prokaryotic adaptive immune systems generally target DNA substrates, type III and VI CRISPR systems direct interference complexes against single-stranded RNA substrates. In type VI systems, the single-subunit C2c2 protein functions as an RNA-guided RNA endonuclease (RNase). How this enzyme acquires mature CRISPR RNAs (crRNAs) that are essential for immune surveillance and how it carries out crRNA-mediated RNA cleavage remain unclear. Here we show that bacterial C2c2 possesses a unique RNase activity responsible for CRISPR RNA maturation that is distinct from its RNA-activated single-stranded RNA degradation activity. These dual RNase functions are chemically and mechanistically different from each other and from the crRNA-processing behaviour of the evolutionarily unrelated CRISPR enzyme Cpf1 (ref. 11). The two RNase activities of C2c2 enable multiplexed processing and loading of guide RNAs that in turn allow sensitive detection of cellular transcripts.
Figures
Extended Data Figure 1. Complete phylogenetic tree of C2c2 family and C2c2 alignment
a, Maximum-likelihood phylogenetic reconstuction of C2c2 proteins. Leaves include GI protein numbers and organism of origin; bootstrap support values, out of 100 resamplings, are presented for inner split. Scale is in substitutions per site. b, Multiple sequence alignment of the three analyzed homologs of C2c2; coordinates are based on LbuC2c2.
Extended Data Figure 2. Purification and Production of C2c2
All C2c2 homologs were expressed in E. coli as His-MBP fusions and purified by a combination of affinity, ion exchange and size exclusion chromatography. The Ni+ affinity tag was removed by incubation with TEV protease. Representative SDS-PAGE gels of chromatography fractions are shown in (a, b). c, The chromatogram from Superdex 200 (16/60) column demonstrating that C2c2 elutes as a single peak, devoid of nucleic acid. d, SDS PAGE analysis of purified proteins used in this manuscript.
Extended Data Figure 3. Mapping of pre-crRNA processing by C2c2 in vitro and in vivo.
a, Cleavage site mapping of LseC2c2 and LshCc2c2 cleavage of a single cognate pre-crRNA array. OH: alkaline hydrolysis ladder; T1: T1 RNase hydrolysis ladder. Cleavage reactions were performed with 100 nM C2c2 and <1 nM pre-crRNA. b–i,Re-analysis of LshC2c2 (b–f) and LseC2c2 (g–i) CRISPR array RNA sequencing experiments from Shmakov et al. (Fig. S7 and Fig. 5, respectively). All reads (b,g) and filtered reads (55 nt or less; as per original Shmakov et al. analysis; c,h) were stringently aligned to each CRISPR array using Bowtie2 (see Methods). Detailed views of individual CRISPR repeat-spacers are shown for Lsh (d–f) and Lse (i). Differences in 5′ end pre-crRNA processing are indicated by arrows below each sequence. BAM alignment files of our analysis are available in Supplementary Materials. This mapping clearly indicates that the 5′ ends of small RNA sequencing reads generated from Lsh pre-crRNAs map to a position 2 nts from the base of the predicted hairpin, in agreement with our_in vitro_ processing data (a). This pattern holds for all mature crRNAs detected from both native expression in_L. shahii_ and heterologous expression in E. coli (data not shown, BAM file available in supplementary methods). Unfortunately, the LseC2c2 crRNA sequencing data (used ing–i) is less informative due to low read depth, and each aligned crRNA exhibits a slightly different 5′ end with little obvious uniformity. The mapping for one of the processed repeats (repeat-spacer 2; i) is in agreement with our data but only with low confidence due to the insufficient read depth.
Extended Data Figure 4. Pre-crRNA processing by C2c2 is spacer-sequence independent, can occur on tandem crRNA arrays, is affected by mutations in the 5′ flanking region of the pre-cRNA and produces a 3′ phosphate product
a, Cleavage site mapping of LbuCc2c2 cleavage of a tandem pre-crRNA array. OH: alkaline hydrolysis ladder; T1: T1 RNase hydrolysis ladder. Cleavage reactions were performed with 100 nM LbuC2c2 and <1 nM pre-crRNA. A schematic of cleavage products is depicted on right, with arrows indicating the mapped C2c2 cleavage products. b, LbuC2c2 4-mer mutant pre-crRNA processing data demonstrating the importance of the 5′ single-stranded flanking region for efficient pre-crRNA processing. Percentage of pre-crRNA processing was measured after 60 min (mean ± s.d., n = 3). **c,**Representative LbuC2c2 pre-crRNA cleavage time-course demonstrating that similar rates of pre-crRNA processing occur independent of crRNA spacer sequence pseudo-first-order rate constants (_k_obs) (mean ± s.d.) are 0.07 ± 0.04 min−1 and 0.08 ± 0.04 min−1 for spacer A and spacer λ2, respectively. d, End group analysis of cleaved RNA by T4 polynucleotide kinase (PNK) treatment. Standard processing assay conditions were used to generate cleavage product, which was then incubated with PNK for 1 hr to remove any 2′, 3′-cyclic phosphates/3′ monophosphates. Retarded migration of band indicates removal of the charged, monophosphate from the 3′ end of radiolabeled 5′ product.
Extended Data Figure 5. LbuC2c2 catalyzes guide-dependent ssRNA degradation on cis and trans targets
a, Schematic of the two modes of C2c2, guide-dependent ssRNA degradation. b, Cleavage of two distinct radiolabeled ssRNA substrates, A and B, by LbuC2c2. Complexes of 100 nM C2c2 and 50 nM crRNA were pre-formed at 37 °C, and reaction was initiated upon addition of <1 nM 5′-labeled target RNA at 25°C.Trans cleavage reactions contained equimolar (<1 nM) concentrations of radiolabeled non-guide-complementary substrate, and unlabeled on-target ssRNA. For multiple ssRNA substrates, we observed that LbuC2c2 catalyzed efficient cleavage only when bound to the complementary crRNA, indicating that LbuC2c2:crRNA cleaves ssRNA in an RNA-guided fashion This activity is hereafter referred to as on-target or_cis-_target cleavage. LbuC2c2-mediated _cis_cleavage resulted in a laddering of multiple products, with cleavage preferentially occurring before uracil residues, analogous to LshC2c2. We repeated non-target cleavage reactions in the presence of unlabeled, on-target (crRNA-complementary) ssRNA. In contrast to non-target cleavage experiments performed in cis, we observed rapid degradation of non-target RNA in trans. The similar RNA cleavage rates and near identical cleavage products observed for both cis_on-target cleavage and trans non-target cleavage implicate the same nuclease center in both activities. c, LbuC2c2 loaded with crRNA targeting spacer A was tested for cleavage activity under both_cis (target A labeled) and trans(target B labeled in the presence of unlabeled target A) cleavage conditions in the presence of 25 mM EDTA.
Extended Data Figure 6. LbuC2c2 ssRNA target cleavage site mapping
a, ssRNA target cleavage assay conducted per Methods demonstrating LbuC2c2-mediated ‘_cis’-cleavage of several radiolabeled ssRNA substrates with identical spacer-complementary sequences but distinct 5′ flanking sequences of variable length and nucleotide composition. Sequences of ssRNA substrates are shown to the right with spacer-complementary sequences for crRNA-A highlighted in yellow. Arrows indicate detected cleavage sites. Gel was cropped for clarity. It should be noted that the pattern of cleavage products produced on different substrates (e.g. A.1 vs. A.2 vs. A.3) indicates that the cleavage site choice is primarily driven by a uracil preference and exhibits an apparent lack of exclusive cleavage mechanism within the crRNA-complementary target sequence, which is in contrast to what is observed for other Class II CRISPR single effector complexes such as Cas9 and Cpf1,. Interestingly, the cleavage pattern observed for substrate A.0 hints at a secondary preference for polyG sequences. b, LbuC2c2 ssRNA target cleavage assay as per Methods, using a range of crRNAs that tile the length of the ssRNA target. The sequence of the ssRNA substrates used in this experiment is shown below the gel with spacer-complementary sequences for each crRNA highlighted in yellow. Arrows indicate predicted cleavage sites. Above each set of lanes, a small diagram indicates the location of the spacer sequence along the target (yellow box) and the cleavage products observed (red arrows) or absent (black arrows). Likewise, it should be noted that for every crRNA the cleavage product length distribution is very similar, again indicating an apparent lack of exclusive cleavage within the crRNA-bound sequence. The absence of a several cleavage products in a subset of the reactions might be explained by the presence of bound C2c2:crRNA on the ssRNA target, which could sterically occlude access to uracils by any_cis (intramolecular) or trans(intermolecular) LbuC2c2 active sites. While proper analysis for protospacer flanking site (PFS) preference for LbuC2c2 is beyond the scope of this study, minimal impact of the 3′ flanking nucleotide was observed. Expected PFS base is noted in diagram next to each guide tested in red.
Extended Data Figure 7. Dependence of RNA targeting on crRNA variants, temperature and point mutations
a, LbuC2c2 ssRNA target cleavage assay carried out, as per Methods with crRNAs possessing 16-nt, 20-nt or 24-nt spacers.b, LbuC2c2 ssRNA target cleavage time-course carried out at either 25°C and 37°C as per methods. c, LbuC2c2 ssRNA target cleavage timecourse carried out as per Methods with crRNAs possessing different 5′-flanking nucleotide mutations. Mutations are highlighted in red. 1–2 nucleotide 5′ extensions negligibly impacted cleavage efficiencies. In contrast, shortening the flanking region to 3 nts slowed cleavage rates. d Impact of point mutations on ribonuclease activity of C2c2 in conserved residue mutants within HEPN motifs for ssRNA targeting.
Extended Data Figure 8. Binding data for LbuC2c2 to mature crRNA and target ssRNA
a, Filter binding assays were conducted as described in the Methods to determine the binding affinity of mature crRNA-A_GG to LbuC2c2-WT, LbuC2c2-dHEPN1, LbuC2c2-dHEPN2, or LbuC2c2-dHEPN1/dHEPN2. The quantified data were fit to standard binding isotherms. Error bars represent the standard deviation from three independent experiments. Measured dissociation constants from three independent experiments (mean ± sd) were 27.1 ± 7.5 nM (LbuC2c2-WT), 15.2 ± 3.2 nM (LbuC2c2-dHEPN1), 11.5 ± 2.5 nM (LbuC2c2-dHEPN2), and 43.3 ± 11.5 nM (LbuC2c2- dHEPN1/dHEPN2). b, Representative electrophoretic mobility shift assay for binding reactions between LbuC2c2-dHEPN1/dHEPN2: crRNA-A_GG and either ‘on-target’ A ssRNA or ‘off-target’ B ssRNA, as indicated. Three independent experiments were conducted as described in the Methods. The gel was cropped for clarity. c, Quantified binding data from (b) were fitted to standard binding isoforms. Error bars represent the standard deviation from three independent experiments. Measured dissociation constants from three independent experiments (mean ± sd) were 1.62 ± 0.43 nM for ssRNA A and N.D (≫10 nM) for ssRNA B.d, Filter binding assays were conducted as described in the Methods to determine the binding affinity of mature crRNA-A_GA to LbuC2c2-WT and LbuC2c2-R1079A. The quantified data were fit to standard binding isotherms. Error bars represent the standard deviation from three independent experiments. Measured dissociation constants from three independent experiments (mean ± sd) were 4.65 ± 0.6 nM (LbuC2c2-WT) and 2.52 ± 0.5 nM (LbuC2c2-R1079A). It is of note that these binding affinities differ from panel a. This difference is accounted for in a slight difference in the 5′ sequence of the guide with panel a guides beginning with a 5′-GGCCA… and panel d 5′-GACCA. While the native sequence guide (5′-GACCA) binds tighter to LbuC2c2, no difference is seen in the RNA targeting efficiencies of these guide variants (Extended Data Fig. 6c).
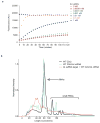
Extended Data Figure 9. RNase detection assay λ2-ssRNA time-course
a, LbuC2c2:crRNA-λ2 was incubated with RNAase-Alert substrate (Thermo-Fisher)) and 100 ng HeLa total RNA in the presence of increasing amounts of λ2 ssRNA (0–1 nM) for 120 min at 37°C. Fluorescence measurements were taken every 5 min. The 1 nM λ2 ssRNA reaction reached saturation before the first time point could be measured. Error bars represent the standard deviation from three independent experiments. **b,**LbuC2c2:crRNA-λ4 or apo LbuC2c2 was incubated in HeLa total RNA for 2 hours in the presence or absence of on-target activating λ4 ssRNA. Degradation of background small RNA was resolved on a small RNA chip in a Bioanalyzer 2100 as per Methods. Small differences are seen in the fragment profile of between apo LbuC2c2 and LbuC2c2:crRNA-λ4. In contrast, upon addition of the on-target ssRNA to the reaction, a drastic broadening and shifting of the tRNA peak reveals extensive degradation of other structured and nonstructured RNA’s present in the reaction upon activation of LbuC2c2 trans activity.
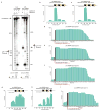
Figure 1. C2c2 proteins process precursor crRNA transcripts to generate mature crRNAs
a, Maximum-likelihood phylogenetic tree of C2c2 proteins. Homologs used in this study are highlighted in yellow. b, Diagram of the three Type VI CRISPR loci used in this study. Black rectangles denote repeat elements, yellow diamonds denote spacer sequences. Cas1 and Cas2 are only found in the genomic vicinity of LshC2c2. c, C2c2-mediated cleavage of pre-crRNA derived from the LbuC2c2, LseC2c2 and LshC2c2 CRISPR repeat loci. OH: alkaline hydrolysis ladder; T1: RNase T1 hydrolysis ladder; processing cleavage reactions were performed with 100 nM C2c2 and <1 nM pre-crRNA. Schematic of cleavage is depicted on right, and predicted pre-crRNA secondary structures are diagrammed below, with arrows indicating the mapped C2c2 cleavage sites.
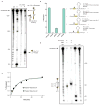
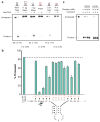
Figure 2. LbuC2c2 mediated crRNA biogenesis depends on both structure and sequence of CRISPR repeats
a, Representative cleavage assay by LbuC2c2 on pre-crRNAs containing structural mutations within the stem and loop regions of hairpin. Processed percentages listed below are quantified at 60 min (mean ± s.d., n = 3). b, Bar graph showing the dependence of pre-crRNA processing on the CRISPR repeat sequence. The wild-type repeat sequence is shown below with individual bars representing tandem nucleotide mutations as noted in red. The cleavage site is indicated by cartoon scissors. Percentage processed was measured after 60 min (mean ± s.d., n = 3). Diagrammed hairpins of tested mutants can be found in Extended Data Figs. 3–4c, Divalent metal ion dependence of the crRNA processing reaction was tested by addition of 10–50 mM EDTA and EGTA to standard reaction conditions.
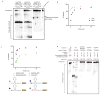
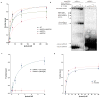
Figure 3. LbuC2c2 contains two distinct ribonuclease activities
a, Quantified time-course data of cis ssRNA target (black) and pre-crRNA (teal) cleavage by LbuC2c2 performed at 37°C. Exponential fits are shown as solid lines (n=3), and the calculated pseudo-first-order rate constants (_k_obs) (mean ± s.d.) are 9.74 ± 1.15 min−1 and 0.12 ± 0.02 min−1 for cis ssRNA target and pre-crRNA cleavage, respectively. b, LbuC2c2 architecture depicting the location of HEPN motifs and processing deficient point mutantc,d Ribonuclease activity of LbuC2c2 mutants for pre-crRNA processing in c and ssRNA targeting in d and Extended Data Fig 6d.
Figure 4. C2c2 provides sensitive detection of transcripts in complex mixtures
a, Illustration of LbuC2c2 RNA detection approach using a quenched fluorescent RNA reporter. b, Quantification of fluorescence signal generated by LbuC2c2 after 30 min for varying concentrations of target RNA in the presence of human total RNA. RNase A shown as positive RNA degradation control. (mean ± s.d., n = 3) c,. Quantification of fluorescence signal generated by LbuC2c2 loaded with a_β_-actin targeting crRNA after 3h for varying amounts of human total RNA or bacterial total RNA (as a_β_-actin null negative control). (mean ± s.d., n = 3) d, Tandem pre-crRNA processing also enables RNA detection. (mean ± s.d., n = 3) e, Model of the Type VI CRISPR pathway highlighting both of C2c2’s ribonuclease activities.
Similar articles
- The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. Fonfara I, et al. Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362 - Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems.
O'Connell MR. O'Connell MR. J Mol Biol. 2019 Jan 4;431(1):66-87. doi: 10.1016/j.jmb.2018.06.029. Epub 2018 Jun 22. J Mol Biol. 2019. PMID: 29940185 Review. - RNA Targeting by Functionally Orthogonal Type VI-A CRISPR-Cas Enzymes.
East-Seletsky A, O'Connell MR, Burstein D, Knott GJ, Doudna JA. East-Seletsky A, et al. Mol Cell. 2017 May 4;66(3):373-383.e3. doi: 10.1016/j.molcel.2017.04.008. Mol Cell. 2017. PMID: 28475872 Free PMC article. - C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector.
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. Abudayyeh OO, et al. Science. 2016 Aug 5;353(6299):aaf5573. doi: 10.1126/science.aaf5573. Epub 2016 Jun 2. Science. 2016. PMID: 27256883 Free PMC article. - Approaches to study CRISPR RNA biogenesis and the key players involved.
Behler J, Hess WR. Behler J, et al. Methods. 2020 Feb 1;172:12-26. doi: 10.1016/j.ymeth.2019.07.015. Epub 2019 Jul 17. Methods. 2020. PMID: 31325492 Review.
Cited by
- Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations.
Liu Q, Guo X, Xun G, Li Z, Chong Y, Yang L, Wang H, Zhang F, Luo S, Cui L, Zhao P, Ye X, Xu H, Lu H, Li X, Deng Z, Li K, Feng Y. Liu Q, et al. Nucleic Acids Res. 2021 Jul 21;49(13):e75. doi: 10.1093/nar/gkab274. Nucleic Acids Res. 2021. PMID: 33905513 Free PMC article. - Recent advances in the use of the CRISPR-Cas system for the detection of infectious pathogens.
Gao H, Shang Z, Chan SY, Ma D. Gao H, et al. J Zhejiang Univ Sci B. 2022 Nov 15;23(11):881-898. doi: 10.1631/jzus.B2200068. J Zhejiang Univ Sci B. 2022. PMID: 36379609 Free PMC article. Review. - CRISPR-based functional genomics for neurological disease.
Kampmann M. Kampmann M. Nat Rev Neurol. 2020 Sep;16(9):465-480. doi: 10.1038/s41582-020-0373-z. Epub 2020 Jul 8. Nat Rev Neurol. 2020. PMID: 32641861 Free PMC article. Review. - Applying CRISPR-Cas9 screens to dissect hematological malignancies.
Iyer DN, Schimmer AD, Chang H. Iyer DN, et al. Blood Adv. 2023 May 23;7(10):2252-2270. doi: 10.1182/bloodadvances.2022008966. Blood Adv. 2023. PMID: 36355853 Free PMC article. Review. - Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Cui N, Zhang JT, Li Z, Liu XY, Wang C, Huang H, Jia N. Cui N, et al. Nat Commun. 2022 Dec 7;13(1):7549. doi: 10.1038/s41467-022-35275-5. Nat Commun. 2022. PMID: 36477448 Free PMC article.
References
- Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. - PubMed
- Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials