The GATOR1 Complex Regulates Metabolic Homeostasis and the Response to Nutrient Stress in Drosophila melanogaster - PubMed (original) (raw)
The GATOR1 Complex Regulates Metabolic Homeostasis and the Response to Nutrient Stress in Drosophila melanogaster
Youheng Wei et al. G3 (Bethesda). 2016.
Abstract
TORC1 regulates metabolism and growth in response to a large array of upstream inputs. The evolutionarily conserved trimeric GATOR1 complex inhibits TORC1 activity in response to amino acid limitation. In humans, the GATOR1 complex has been implicated in a wide array of pathologies including cancer and hereditary forms of epilepsy. However, the precise role of GATOR1 in animal physiology remains largely undefined. Here, we characterize null mutants of the GATOR1 components nprl2, nprl3, and iml1 in Drosophila melanogaster We demonstrate that all three mutants have inappropriately high baseline levels of TORC1 activity and decreased adult viability. Consistent with increased TORC1 activity, GATOR1 mutants exhibit a cell autonomous increase in cell growth. Notably, escaper nprl2 and nprl3 mutant adults have a profound locomotion defect. In line with a nonautonomous role in the regulation of systemic metabolism, expressing the Nprl3 protein in the fat body, a nutrient storage organ, and hemocytes but not muscles and neurons rescues the motility of nprl3 mutants. Finally, we show that nprl2 and nprl3 mutants fail to activate autophagy in response to amino acid limitation and are extremely sensitive to both amino acid and complete starvation. Thus, in Drosophila, in addition to maintaining baseline levels of TORC1 activity, the GATOR1 complex has retained a critical role in the response to nutrient stress. In summary, the TORC1 inhibitor GATOR1 contributes to multiple aspects of the development and physiology of Drosophila.
Keywords: Iml1; Nprl2; Nprl3; TORC1; metabolism.
Copyright © 2016 Wei et al.
Figures
Figure 1
Schematic representation of GATOR1 mutants generated using CRISPR/Cas9 genome editing. To generate the nprl2 and iml1 deletion mutants, two gRNAs that target the 5′ and 3′ of each gene were used. (A) nprl2 encodes a transcript of 1707 bp and a protein of 412 amino acids (FBgn0030800). The nprl21 deletion contains 30 amino acids from the N-terminus and four frame-shifted amino acids from the C-terminus. (B) iml1 encodes five transcripts (FBgn0035227). These transcripts encode proteins of 1471, 1472, 1503, 1511, and 1544 amino acids, respectively. The iml11 deletion contains 17 amino acids from the N-terminus and 53 amino acids from the C-terminus of all iml1 transcripts. Notably, the intron of iml1 also encodes a transcript Ir62a, which is also deleted. CDS, coding sequences; CRISPR, clustered regularly interspaced short palindromic repeats; gRNA, guide RNA; UTR, untranslated region.
Figure 2
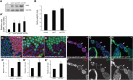
GATOR1 is required to maintain baseline levels of TORC1 activity in Drosophila. (A) Third star larvae were lysed in RIPA buffer. The protein levels of phospho-T398-S6K and total S6K were determined by western blot. (A’) The ratio of pS6K/S6K in the y, w lysis was set as 1. Error bars indicate SD of three independent experiments. * P < 0.05, ** P < 0.01. (B) The body weight of male flies. Error bars indicate SD of five independent experiments. ** P < 0.01. (C–E) Ovaries were dissected and stained with DAPI and anti-1B1 antibody. The nprl21, nprl31, and iml1 1 homozygous follicles cell clones were identified by the absence of RFP or GFP. The 1B1 antibody stains cell membranes and was used to mark the outline of the cells (C’–E’). The area of homozygous cells and adjacent wild-type cells were quantified using Image J. The mean value of the wild-type cell size was set as 1 for each image. For each genotype, eight images were quantified. Error bars indicate SD. ** P < 0.01. (F–H) Ovaries were dissected and stained with DAPI. The nprl21, nprl31, and iml1 1 homozygous egg chamber clones were identified by the absence of RFP and GFP. Homozygous mutant cells are indicated by an arrow, while older wild-type cells are indicated by an arrowhead. Note that younger homozygous mutant cells are larger than older wild-type cells. (F’–H’) In order to clearly show the clonal boundary, RFP and GFP channels are shown separately. DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; RFP, red fluorescent protein; RIPA, radioimmunoprecipitation assay.
Figure 3
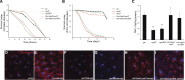
nprl2 and nprl3 mutants are sensitive to starvation. (A) Newly hatched male flies were cultured at 25° on standard food for 3 d, and then transferred to starvation media (0.8% agar in PBS) and counted at the indicated time points. The survival curves of wild-type, nprl21, nprl31/Df, Ubi-GAL4/UAS-Nprl2; nprl21, and Ubi-GAL4/UAS-Nprl3; nprl31/Df are shown. (B) Newly hatched male flies were cultured at 25° on standard food for 3 d, and then transferred to amino acid starvation media (20% sucrose, 0.8% agar in PBS) and counted each day. The survival curves of wild-type, nprl21, nprl31/Df, Ubi-GAL4/UAS-Nprl2; nprl21, and Ubi-GAL4/UAS-Nprl3; nprl31/Df are shown. (C) Newly hatched male flies were cultured at 25° on standard food for 3 d and then lysed. Total body TAG and protein were measured. Protein levels were used for normalization. The TAG to protein ratio is shown. Error bars indicate SD of three independent experiments. ** P < 0.01. (D–I) Third instar wild-type_, nprl21, nprl31/Df, Ubi-GAL4/UAS-Nprl2; nprl21, and _Da>-GAL4/UAS-Nprl3; nprl31/Df larvae were fed or starved for 4 hr. Subsequently, fat bodies were removed and stained with Hoechst and LysoTracker Red. PBS, phosphate-buffered saline; TAG, triglycerides.
Figure 4
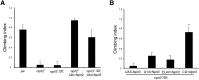
Nprl2 and Nprl3 are required in the fat body for Drosophila locomotion. (A) The climbing indices of the indicated genotypes are shown. Overexpression of Nprl2 and Nprl3 using the ubiquitous Ubi-GAL4 driver suppressed the climbing defects of nprl21 and nprl31/Df mutant flies. Error bars indicate SD of at least three independent experiments. ** P < 0.01. (B) The climbing indices of nprl31/Df mutant flies that overexpress the Nprl3 protein using a neuronal driver ELAV-GAL4, a muscle driver G14-GAL4, or a fat body, hemocyte driver CG-GAL4 are shown. Error bars indicate SD of at least six independent experiments. ** P < 0.01.
Similar articles
- The GATOR2 Component Wdr24 Regulates TORC1 Activity and Lysosome Function.
Cai W, Wei Y, Jarnik M, Reich J, Lilly MA. Cai W, et al. PLoS Genet. 2016 May 11;12(5):e1006036. doi: 10.1371/journal.pgen.1006036. eCollection 2016 May. PLoS Genet. 2016. PMID: 27166823 Free PMC article. - TORC1 regulators Iml1/GATOR1 and GATOR2 control meiotic entry and oocyte development in Drosophila.
Wei Y, Reveal B, Reich J, Laursen WJ, Senger S, Akbar T, Iida-Jones T, Cai W, Jarnik M, Lilly MA. Wei Y, et al. Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):E5670-7. doi: 10.1073/pnas.1419156112. Epub 2014 Dec 15. Proc Natl Acad Sci U S A. 2014. PMID: 25512509 Free PMC article. - The TORC1 inhibitors Nprl2 and Nprl3 mediate an adaptive response to amino-acid starvation in Drosophila.
Wei Y, Lilly MA. Wei Y, et al. Cell Death Differ. 2014 Sep;21(9):1460-8. doi: 10.1038/cdd.2014.63. Epub 2014 May 2. Cell Death Differ. 2014. PMID: 24786828 Free PMC article. - GATOR1 complex: the common genetic actor in focal epilepsies.
Baldassari S, Licchetta L, Tinuper P, Bisulli F, Pippucci T. Baldassari S, et al. J Med Genet. 2016 Aug;53(8):503-10. doi: 10.1136/jmedgenet-2016-103883. Epub 2016 May 19. J Med Genet. 2016. PMID: 27208208 Review. - Evolutionarily conserved regulation of TOR signalling.
Takahara T, Maeda T. Takahara T, et al. J Biochem. 2013 Jul;154(1):1-10. doi: 10.1093/jb/mvt047. Epub 2013 May 21. J Biochem. 2013. PMID: 23698095 Review.
Cited by
- Unveiling GATOR2 Function: Novel Insights from Drosophila Research.
Bettedi L, Zhang Y, Yang S, Lilly MA. Bettedi L, et al. Cells. 2024 Oct 30;13(21):1795. doi: 10.3390/cells13211795. Cells. 2024. PMID: 39513902 Free PMC article. Review. - SEA and GATOR 10 Years Later.
Loissell-Baltazar YA, Dokudovskaya S. Loissell-Baltazar YA, et al. Cells. 2021 Oct 8;10(10):2689. doi: 10.3390/cells10102689. Cells. 2021. PMID: 34685669 Free PMC article. Review. - Royal jelly attenuates metabolic defects in a Drosophila mutant with elevated TORC1 activity.
Cheng Y, Cai J, Fu Y, Feng C, Hao Y, Wei Y. Cheng Y, et al. Biol Open. 2020 Nov 6;9(11):bio054999. doi: 10.1242/bio.054999. Biol Open. 2020. PMID: 33037015 Free PMC article. - Differential amino acid transporter expression in adult Drosophila melanogaster tissues.
Wright Y, Armstrong AR. Wright Y, et al. MicroPubl Biol. 2024 Feb 27;2024:10.17912/micropub.biology.001121. doi: 10.17912/micropub.biology.001121. eCollection 2024. MicroPubl Biol. 2024. PMID: 38481556 Free PMC article. - FKBP39 controls nutrient dependent Nprl3 expression and TORC1 activity in Drosophila.
Zhou Y, Guo J, Wang X, Cheng Y, Guan J, Barman P, Sun MA, Fu Y, Wei W, Feng C, Lilly MA, Wei Y. Zhou Y, et al. Cell Death Dis. 2021 Jun 2;12(6):571. doi: 10.1038/s41419-021-03860-z. Cell Death Dis. 2021. PMID: 34078879 Free PMC article.
References
- Aberle H., Haghighi A. P., Fetter R. D., McCabe B. D., Magalhaes T. R., et al. , 2002. Wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron 33: 545–558. - PubMed
- Bainbridge S. P., Bownes M., 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66: 57–80. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases