Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening - PubMed (original) (raw)
. 2016 Nov 8;7(45):72593-72607.
doi: 10.18632/oncotarget.12241.
Salar Khaleghzadegan 1, Brian Mears 1, Koji Hatano 1, Tarana A Kudrolli 1, Wasim H Chowdhury 1, David B Yeater 1, Charles M Ewing 1, Jun Luo 1, William B Isaacs 1 2, Luigi Marchionni 2, Shawn E Lupold 1 2
Affiliations
- PMID: 27683042
- PMCID: PMC5341930
- DOI: 10.18632/oncotarget.12241
Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening
Binod Kumar et al. Oncotarget. 2016.
Abstract
The Androgen Receptor (AR) plays a key role in prostate biology and in the progression of prostate cancer (PCa) to castration resistance. The role of microRNAs (miRNAs) in aberrant AR signaling have not been fully characterized. Here we screened a library of 810 miRNA mimics to identify miRNAs that alter AR activity in complementary functional assays including protein lysate microarray (LMA) quantification of AR and PSA protein levels, AR transcriptional reporter activity, and AR-positive PCa cell viability. Candidate AR-regulating miRNAs were verified through AR transcriptional reporter and cell viability assays. MiRNA binding sites were found within the AR 3'-untranslated region (UTR) and within the AR and AR-V7 coding regions. MiRNA activity was characterized by western blotting, 3'-UTR reporter assay, and AR-GFP and AR-V7-GFP reporter assays. Results uncovered miR-30 family members as direct AR inhibitors. Inhibition of endogenous miR-30b-3p and miR-30d-5p enhanced AR expression and androgen-independent cell growth. Droplet digital RT-PCR quantification of miR-30c-5p and miR-30d-5p revealed significantly reduced levels in metastatic castration resistant PCa (CRPC), when compared to healthy prostate tissues. MiR-30d-5p levels were inversely correlated with AR activity, as measured by PSA mRNA, in metastatic CRPC. Collectively, these studies provide a comprehensive evaluation of AR-regulating miRNAs in PCa.
Keywords: miR-30; miRNA; microRNA; androgen receptor; prostate cancer; castration resistant prostate cancer.
Conflict of interest statement
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Figures
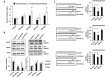
Figure 1. High-throughput miRNA mimic library screen for miRNAs that modulate the AR Signaling Axis
A, Schematic of miRNA mimic screen for AR and PSA protein level by LMA. B, Log transformed FC in AR protein level, relative to control miRNA, in LNCaP cells 48 hours post transfection. AR, normalized to GAPDH, is organized as a waterfall plot for each mimic, ranked by FC. C, Log transformed FC in PSA protein level, relative to control miRNA, in LNCaP cells 48 hours post transfection. D, Schematic of miRNA mimic screens for AR transcriptional activity and cell viability. AR was stimulated 48 hours post transfection by 5 nM R1881, and activity detected 24 hours later. Viable cell density was quantified by MLuc Viability Assay 6 days post transfection. E, Log transformed FC in AR transcriptional activity as measured by PSE-PBN-Luc reporter, relative to control miRNA and normalized to Renilla luciferase, in LNCaP cells. F, Log transformed FC in LNCaP-MLuc viable cell density as measured by MLuc Cell Viability Assay, relative to control miRNA, 6 days post transfection. Waterfall plots: Dashed line indicates negative control mimic. Percentage of miRNAs with signal above and below control miRNA are indicated.
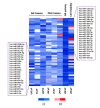
Figure 2. Candidate AR regulating miRNA mimics
48 candidate mimics. The top 25% of inhibitory miRNA mimics from each screen and cell line were compared to identify 43 candidates which inhibited AR transcriptional activity, as well as AR and PSA protein level, in at least two cell lines. The five most potent inhibitors of AR transcription activity were also included. For visualization, a heat map of linear FC in signal for each mimic, assay, and cell line is presented with red indicating an increase in signal and blue indicating a decrease in signal.
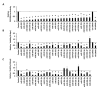
Figure 3. Verification of candidate miRNAs by AR Transcriptional Activity analysis and effects of verified mimics on cell viability
A, AR transcriptional activity of the top 25 verified candidates as measured by PSE-PBN-Luc reporter, relative to control miRNA and normalized to Renilla luciferase, in LNCaP cells. AR was stimulated 48 hours post transfection by 5 nM R1881, and activity detected 24 hours later. B, Relative viability of LNCaP-MLuc cells, as measured by MLuc Cell Viability Assay, 6 days post transfection with the top 25 verified candidates. C, Relative viability of PC3-MLuc cells, as measured by MLuc Cell Viability Assay, 6 days post transfection with the top 25 verified candidates. Graphs represents average ± Standard Error (SE) from at least two biological replicates performed in triplicate. Dashed line represent control miRNA treated cell activity or viability. Cnt-siRNA, control siRNA, AR-siRNA. Asterisk indicates significant suppression of transcriptional activity or viability. *, p < 0.05.
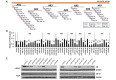
Figure 4. Regulation of AR expression by 3′UTR-targeting miRNAs
A, Map of the extended AR 3′UTR (ENST00000396044.7) and corresponding AR 3′UTR reporter amplicons AR1-AR7. MiRNA binding site and type are indicated by color according to legend. B, AR 3′UTR luciferase activity for each amplicon (AR1-AR7), relative to control miRNA mimic and normalized to Renilla luciferase, 48 hours post miRNA mimic transfection. Data represents average ± SE, from at least two biologic replicates for each miRNA in triplicate. Dashed line indicates control miRNA. Asterisk indicates significant miRNA suppression of luciferase expression, * p < 0.05. C, Western blot analysis for AR (AR) in LNCaP cells, AR-V7 in VCaP cells, and GAPDH 48 hours post mimic transfection (20 nM).
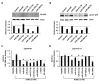
Figure 5. Regulation of AR by miRNAs targeting the coding region
A, PC3 cells were co-transfection with pEGFP-AR and miRNA mimics predicted to bind sites within the AR coding region. AR-GFP was detected by western blot 48 hours post transfection. Bar graphs represent β-actin-normalized intensity, relative to control, from two biologic replicates. B, PC3 anti-GFP western blot 48 hours after co-transfection with pEGFP-AR-V7 and miRNA mimics predicted to bind the AR coding region. Bar graphs represents β-actin-normalized intensity, as above. C, AR transcriptional activity, as measured by Renilla normalized PSE-PBN-Luc activity, after PC3 transfection with pEGFP-AR and mimics. AR was activated 48 hours after transfection with 5 nM R1881 and activity measured 24 hours later. D, AR transcriptional activity, as measured by Renilla normalized PSE-PBN-Luc activity, after PC3 transfection with pEGFP-AR-V7 and mimics. AR activity was stimulated with 5 nM R1881 and activity measured 24 hours later. Bar graphs represent average ± standard deviation (SD) from two biologic replicates. Asterisks indicate significant reduction in AR transcriptional activity, * p < 0.05.
Figure 6. miR-30b-3p and miR-30d-5p directly regulate AR protein expression and PCa cell growth
A, LNCaP-MLuc cells were transfected with control or miR-30 antagomirs. Cell viability was determined 6 days post transfection in complete media (with androgens) or in androgen depleted media by MLuc Cell Viability Assay. Antagomirs: miR-30a-I: miR-30a-3p; miR-30b-I: miR-30b-3p; miR-30c-I: miR-30c-5p; miR-30d-I: miR-30d-5p. Asterisk indicated significant increase in viable cell density, *, p < 0.05. B, AR western blot 48 hours post transfection with control, miR-30 mimics, or miR-30 antagomirs (20 nM) in LNCaP, LAPC4, and VCaP cells. Bar graph represents average GAPDH normalized AR intensity average, relative to control. Error bars = SD. C, AR 3′UTR luciferase activity for amplicons AR1 and AR2, before and after site-directed mutagenesis of miR-30 binding sites. Schematic indicates miRNA seed sequence binding sites and mutations (−M) are indicated by asterisks. Top, miR-30b-3p mimics with AR1-WT and AR1-M in LNCaP; Middle, miR-30b-3p mimics with AR2-WT and AR2-M in LNCaP; Bottom, miR-30d-5p mimics with AR1-WT and AR1-M in LAPC4 cells. Bar graphs represent average 3′UTR activity ± SD, normalized to control miRNA mimics from two biologic replicates. Asterisk indicates significant suppression of 3′UTR activity by miRNA mimics, * p < 0.05.
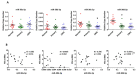
Figure 7. Differential expression of miR-30c-5p and miR-30d-5p in human PCa and correlation with PSA mRNA levels
A, Relative miR-30 expression quantified by ddPCR of reverse transcribed miRNA from normal prostate (N=15); primary prostate cancer (N=15) and metastatic CRPC (N=15). Mean ± SEM. Asterisk indicates significantly reduced miR-30 expression, relative to normal prostate, * p < 0.05. B, Correlation analysis of PSA mRNA level, as a surrogate for AR transcriptional activity, and miR-30 miRNA levels in CRPC samples (N = 15). PSA mRNA level determined by qRT-PCR and normalized to GAPDH. Correlation determined by Pearson correlation calculation. R – Pearson r. Asterisk indicates a significant correlation between PSA mRNA and miR-30, * p < 0.05.
Similar articles
- miR-103a-2-5p/miR-30c-1-3p inhibits the progression of prostate cancer resistance to androgen ablation therapy via targeting androgen receptor variant 7.
Chen W, Yao G, Zhou K. Chen W, et al. J Cell Biochem. 2019 Aug;120(8):14055-14064. doi: 10.1002/jcb.28680. Epub 2019 Apr 8. J Cell Biochem. 2019. PMID: 30963631 - The miRNAs 203a/210-3p/5001-5p regulate the androgen/androgen receptor/YAP-induced migration in prostate cancer cells.
Huo C, Kuo YY, Lin CY, Shiah SG, Li CY, Huang SP, Chen JK, Wang WC, Kung HJ, Chuu CP. Huo C, et al. Cancer Med. 2024 Aug;13(16):e70106. doi: 10.1002/cam4.70106. Cancer Med. 2024. PMID: 39149855 Free PMC article. - Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer.
Coarfa C, Fiskus W, Eedunuri VK, Rajapakshe K, Foley C, Chew SA, Shah SS, Geng C, Shou J, Mohamed JS, O'Malley BW, Mitsiades N. Coarfa C, et al. Oncogene. 2016 May 5;35(18):2345-56. doi: 10.1038/onc.2015.295. Epub 2015 Sep 14. Oncogene. 2016. PMID: 26364608 Free PMC article. - The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer.
Ribas J, Lupold SE. Ribas J, et al. Cell Cycle. 2010 Mar 1;9(5):923-9. doi: 10.4161/cc.9.5.10930. Epub 2010 Mar 11. Cell Cycle. 2010. PMID: 20160498 Free PMC article. Review. - MicroRNA Regulation of Androgen Receptor in Castration-Resistant Prostate Cancer: Premises, Promises, and Potentials.
Ebrahimi S, Hashemy SI, Sahebkar A, Aghaee Bakhtiari SH. Ebrahimi S, et al. Curr Mol Pharmacol. 2021 Oct 25;14(4):559-569. doi: 10.2174/1874467213666201223121850. Curr Mol Pharmacol. 2021. PMID: 33357209 Review.
Cited by
- Extracellular vesicles in prostate cancer: a narrative review.
Hatano K, Fujita K. Hatano K, et al. Transl Androl Urol. 2021 Apr;10(4):1890-1907. doi: 10.21037/tau-20-1210. Transl Androl Urol. 2021. PMID: 33968677 Free PMC article. Review. - MicroRNAs as Guardians of the Prostate: Those Who Stand before Cancer. What Do We Really Know about the Role of microRNAs in Prostate Biology?
Andl T, Ganapathy K, Bossan A, Chakrabarti R. Andl T, et al. Int J Mol Sci. 2020 Jul 7;21(13):4796. doi: 10.3390/ijms21134796. Int J Mol Sci. 2020. PMID: 32645914 Free PMC article. Review. - Clinically Significant Dysregulation of hsa-miR-30d-5p and hsa-let-7b Expression in Patients with Surgically Resected Non-Small Cell Lung Cancer.
Hosseini SM, Soltani BM, Tavallaei M, Mowla SJ, Tafsiri E, Bagheri A, Khorshid HRK. Hosseini SM, et al. Avicenna J Med Biotechnol. 2018 Apr-Jun;10(2):98-104. Avicenna J Med Biotechnol. 2018. PMID: 29849986 Free PMC article. - Deregulated microRNAs Involved in Prostate Cancer Aggressiveness and Treatment Resistance Mechanisms.
Gujrati H, Ha S, Wang BD. Gujrati H, et al. Cancers (Basel). 2023 Jun 10;15(12):3140. doi: 10.3390/cancers15123140. Cancers (Basel). 2023. PMID: 37370750 Free PMC article. Review. - The orally active pterocarpanquinone LQB-118 exhibits cytotoxicity in prostate cancer cell and tumor models through cellular redox stress.
Martino T, Kudrolli TA, Kumar B, Salviano I, Mencalha A, Coelho MGP, Justo G, Costa PRR, Sabino KCC, Lupold SE. Martino T, et al. Prostate. 2018 Feb;78(2):140-151. doi: 10.1002/pros.23455. Epub 2017 Nov 6. Prostate. 2018. PMID: 29105806 Free PMC article.
References
- Brown TR, Lubahn DB, Wilson EM, Joseph DR, French FS, Migeon CJ. Deletion of the steroid-binding domain of the human androgen receptor gene in one family with complete androgen insensitivity syndrome: evidence for further genetic heterogeneity in this syndrome. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(21):8151–8155. - PMC - PubMed
- Huggins C, Hodges CV. Studies on prostatic cancer - I The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer research. 1941;1(4):293–297. - PubMed
- Sharifi N, Gulley JL, Dahut WL. Androgen deprivation therapy for prostate cancer. Jama. 2005;294(2):238–244. - PubMed
- Wong YN, Ferraldeschi R, Attard G, de Bono J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nature reviews Clinical oncology. 2014;11(6):365–376. - PubMed
- Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. The New England journal of medicine. 2004;351(15):1488–1490. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous