Human metapneumovirus Induces Reorganization of the Actin Cytoskeleton for Direct Cell-to-Cell Spread - PubMed (original) (raw)
Human metapneumovirus Induces Reorganization of the Actin Cytoskeleton for Direct Cell-to-Cell Spread
Farah El Najjar et al. PLoS Pathog. 2016.
Abstract
Paramyxovirus spread generally involves assembly of individual viral particles which then infect target cells. We show that infection of human bronchial airway cells with human metapneumovirus (HMPV), a recently identified paramyxovirus which causes significant respiratory disease, results in formation of intercellular extensions and extensive networks of branched cell-associated filaments. Formation of these structures is dependent on actin, but not microtubule, polymerization. Interestingly, using a co-culture assay we show that conditions which block regular infection by HMPV particles, including addition of neutralizing antibodies or removal of cell surface heparan sulfate, did not prevent viral spread from infected to new target cells. In contrast, inhibition of actin polymerization or alterations to Rho GTPase signaling pathways significantly decreased cell-to-cell spread. Furthermore, viral proteins and viral RNA were detected in intercellular extensions, suggesting direct transfer of viral genetic material to new target cells. While roles for paramyxovirus matrix and fusion proteins in membrane deformation have been previously demonstrated, we show that the HMPV phosphoprotein extensively co-localized with actin and induced formation of cellular extensions when transiently expressed, supporting a new model in which a paramyxovirus phosphoprotein is a key player in assembly and spread. Our results reveal a novel mechanism for HMPV direct cell-to-cell spread and provide insights into dissemination of respiratory viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
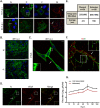
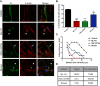
Fig 1. HMPV infection in BEAS-2B cells results in the formation of branched filamentous networks and intercellular extensions.
(A) HMPV infected BEAS-2B cells at 18 or 24 h.p.i., were fixed and processed for immunofluorescence staining. White arrows indicate intercellular extensions, red arrowheads indicate branched viral filaments and white arrowheads indicate viral filaments. Scale bars = 10 μm. (B) HMPV infected BEAS-2B cells at 48 h.p.i. were fixed, processed for immunofluorescence and stained with antibodies for HMPV N or P. Scale bar = 50 μm. (C) BEAS-2B cells were inoculated with PIV5 and at 48 h.p.i. cells were fixed, processed for immunofluorescence and stained with antibody against the F protein of PIV5. Arrow indicates an intercellular extension in a PIV5-infected cell and inset shows filaments. Scale bar = 10 μm. (D) BEAS-2B cells were inoculated with HMPV at M.O.I. of 3, and at 24 h.p.i; cells were fixed and stained with the plasma membrane marker WGA and immunostained for HMPV N. Scale bar = 10 μm. (E) Table showing range and average diameter of filaments and intercellular extensions. Images were taken for a total of 100 HMPV-infected cells and imageJ analysis tool was used to determine the diameter of branched filaments and intercellular extensions. (F) Cells were inoculated with HMPV, and 24h later, cells were processed for Stochastic Optical Reconstruction Microscopy (STORM) and stained with an anti-N antibody (green) and an anti-M antibody (red). Inset shows filaments with a central core of N protein surrounded by matrix protein. Scale bar = 1 μm. (G) BEAS-2B cells were inoculated with rgHMPV at an M.O.I. of 2, and at different times post infection, cells or culture media were collected and virus titers determined. Graph shows mean ± SD for three independent experiments.
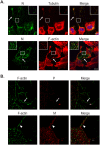
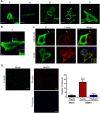
Fig 2. Actin and tubulin are present in intercellular extensions and branched filamentous networks.
(A) BEAS-2B cells were inoculated with HMPV, and 24 h.p.i. cells were fixed and stained for HMPV N, tubulin or F-actin. Inset shows colocalization of actin and tubulin with N in branched filaments and arrows indicate intercellular extensions. Scale bar = 50μm. (B) Cells were infected with HMPV for 24h, processed for Stochastic Optical Reconstruction Microscopy (STORM) and stained with an anti-P or anti-M antibody (red) and phalloidin (green). Arrows show a branched filament and arrowheads show a filament. Scale bar = 1 μM.
Fig 3. Actin and microtubules have different roles in formation of branched filamentous networks and intercellular extensions.
(A) and (B) BEAS-2B cells, were inoculated with HMPV and 2 h.p.i. DMSO vehicle or the indicated drug was added, and cells were incubated for an additional 22 hours. Cells were then fixed and processed for immunofluorescence staining. Arrows indicate branched filamentous networks and arrowheads indicate viral filaments. Scale bars = 10 μm. (C) Cells treated as in A and B were processed for immunofluorescence and images acquired. The NeuronJ plugin of the ImageJ analysis tool was used to manually trace 10 cells for each condition from three independent experiments. Sholl analysis was then performed to evaluate level of branching. (D) Cells were inoculated with HMPV for 2 hours and treated with cytochalasin D. Twenty-four h.p.i. cells were processed for STORM and stained with an anti-N antibody (green) and an anti-M antibody (red). (E) BEAS-2B cells were infected with HMPV, drugs were added at 2 h.p.i. and at 24 h.p.i, cells were fixed and stained with WGA and antibody for N. Scale bars = 10μm. (F) Cells were inoculated with rgHMPV at an M.O.I. of 1 and treated with inhibitors at 2 h.p.i. At 48 h.p.i., cells or culture media were collected and virus titers determined. Graph shows mean ± SD for three independent experiments. Statistical analysis was performed using two-way analysis of variance (ANOVA), (*** = p< 0.0001; n.s. = non-significant).
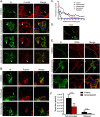
Fig 4. Formation of intercellular extensions requires active actin dynamics.
(A) BEAS-2B cells were mock infected or inoculated with HMPV, and 24 h.p.i. cells were fixed and stained for HMPV N (green) or F-actin (red). Arrows indicate intercellular extensions. (B–D) BEAS-2B cells were either mock infected or infected with HMPV and 24 h.p.i. cells were fixed and processed for immunofluorescence staining. Images were taken, and 80–100 cells from three independent experiments were used to score the presence of extensions (B) or analyzed using the ImageJ analysis tool to determine extension length (C and D). (E) Cells were inoculated with HMPV for 2 hours and incubated with DMSO or the indicated drug, and 24 h.p.i. cells were stained with an antibody recognizing HMPV N (green). Images were acquired on a confocal microscope, and percentage of infected cells with extensions was manually determined for a total of 100 cells from three independent experiments. Scale bars = 50 μm. (F) Images were acquired on a confocal microscope and percentage of infected cells with extensions was manually determined for a total of 100 cells from three independent experiments. Data represent means ± SD. Statistical analysis was performed using one-way analysis of variance (ANOVA), (*** = p< 0.0001; ** = p< 0.001; * = p<0.05).
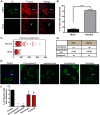
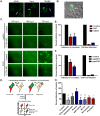
Fig 5. Rho GTPases Cdc42, Rac1 and RhoA contribute to formation of branched filamentous networks and intercellular extensions.
(A) BEAS-2B cells were inoculated with HMPV, and the indicated drug was added 2 hours later. Twenty-four h.p.i. cells were fixed and stained for HMPV N or F-actin. Arrows indicate an intercellular extension. Arrowheads show punctate localization of N, and asterisks indicate short branched filaments. (B) Images acquired from three independent experiments were processed, and a total of 100 infected cells were manually assessed for the presence of extensions. Data represents mean ± SD. Statistical analysis was performed using one-way ANOVA, (*** = p< 0.0001; ** = p< 0.001). (C) A total of 10 cells from three independent experiments were traced using the NeuronJ plugin of the ImageJ analysis tool and the degree of branching was evaluated using Sholl analysis. (D) Table showing the targets of the inhibitors used and their specific concentration.
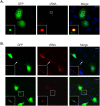
Fig 6. HMPV P induces formation of membrane extensions and co-localizes with actin.
(A) BEAS-2B cells expressing HMPV F, N, M or P were processed for immunofluorescence and stained for the indicated viral protein (green) and DAPI (blue) to stain the nucleus. Insets show membrane extensions in cells transfected with F and M, arrows indicate long membrane extensions and the arrowhead shows a branched filament. (B) A549 cells were transfected with pCAGGS-HMPV P plasmid, and 24 hours post transfection, cells were fixed and processed for immunofluorescence staining. Inset shows membrane ruffling. (C) BEAS-2B or A549 cells were transfected with pCAGGS-HMPV P, and 24 hours later, cells were fixed and stained for P (green) and F-actin (red). Inset shows intracellular colocalization of P and F-actin, and arrows indicate colocalization of P and F-actin in a cellular extension. Scale bars = 10μm. (D) BEAS-2B cells were infected with HMPV, and 24 h.p.i, cells were fixed and proximity ligation assay was performed using the indicated antibodies. Each red signal denotes a reaction due to proximity of P and actin. Scale bars = 50μm. Quantification was done using BlobFinder software analysis tool. Graph shows mean ± SD. Statistical analysis was done using one-way ANOVA (*** = p< 0.0001).
Fig 7. HMPV can spread directly from cell-to-cell.
(A) A549, Vero or 16HBE cells were inoculated with HMPV, and 24 h.p.i., cells were fixed and stained for N protein. Arrows indicate intercellular extensions and arrowhead shows branched filaments. (B) BEAS-2B cells were inoculated with rgHMPV, and 24 h.p.i., cells were prepared for live imaging. Arrow indicates an intercellular extension. (C) BEAS-2B cells were infected with rgHMPV or rgPIV5 at an M.O.I. of 2. Six hours later, infection media was removed and replaced with regular media or media containing 0.75% methylcellulose (MC). Images were taken every 24 hours for 3 days. (D) Schematic of the coculture assay. BEAS-2B cells were inoculated with rgHMPV at an M.O.I. of 2. Forty-eight h.p.i., cells were stained with cell tracker CMRA orange dye for 30 minutes. Infected donor cells were then collected and added to naive target cells at a ratio of 1:1. To study direct cell transmission by blocking re-infection by released particles, the assay was done in the presence of neutralizing antibodies (E) or using target cells that lack a heparan sulfate binding factor for HMPV infection (F). Control represents infection without the presence of neutralizing antibodies (E) or with wt CHO cells (F). Twenty-four hours post coculture, cells were collected and analyzed by flow cytometry. (G) Cells were prepared for coculture assay as described in (D), and the indicated inhibitor was added at the time of mixing donor and infected cells and left in the media for 24 hours. Control represents addition of DMSO only. Data represent means ± SD for three independent experiments. One-way ANOVA was used for statistical analysis (*** = p< 0.0001; ** = p< 0.001; * = p<0.05).
Fig 8. Viral RNA is present in intercellular extensions.
(A) Twenty-four hours post infection with rgHMPV, BEAS-2B cells were fixed, permeabilized with 70% ethanol and incubated with the FISH probes targeting viral RNA overnight. Cells were then washed with 2xSCC buffer and mounted with vectashield. Inset indicates viral RNA in a potential inclusion body. (B) BEAS2B cells infected for 72 hours with rgHMPV were processed as in (A). Arrows indicate an intercellular extension and inset shows larger RNA-containing structure in intercellular extension.
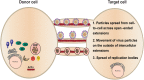
Fig 9. Models for cell-to-cell spread of HMPV in BEAS-2B cells.
Virus particles exit an infected cell and move inside an open-ended intercellular extension to infect a target cell (model 1). In model 2, virus particles move from an infected cell to a target cell at the surface of an intercellular extension. Replication bodies can be transmitted from an infected cell to a target cell along an intercellular extension (model 3).
Similar articles
- Inhibition of Human Metapneumovirus Binding to Heparan Sulfate Blocks Infection in Human Lung Cells and Airway Tissues.
Klimyte EM, Smith SE, Oreste P, Lembo D, Dutch RE. Klimyte EM, et al. J Virol. 2016 Sep 29;90(20):9237-50. doi: 10.1128/JVI.01362-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489270 Free PMC article. - Imaging analysis of human metapneumovirus-infected cells provides evidence for the involvement of F-actin and the raft-lipid microdomains in virus morphogenesis.
Jumat MR, Nguyen Huong T, Wong P, Loo LH, Tan BH, Fenwick F, Toms GL, Sugrue RJ. Jumat MR, et al. Virol J. 2014 Nov 19;11:198. doi: 10.1186/s12985-014-0198-8. Virol J. 2014. PMID: 25408253 Free PMC article. - Imaging analysis reveals budding of filamentous human metapneumovirus virions and direct transfer of inclusion bodies through intercellular extensions.
El Najjar F, Castillo SR, Moncman CL, Wu C-Y, Isla E, Velez Ortega AC, Frolenkov GI, Cifuentes-Munoz N, Dutch RE. El Najjar F, et al. mBio. 2023 Oct 31;14(5):e0158923. doi: 10.1128/mbio.01589-23. Epub 2023 Sep 8. mBio. 2023. PMID: 37681946 Free PMC article. - To assemble or not to assemble: The changing rules of pneumovirus transmission.
Cifuentes-Muñoz N, Ellis Dutch R. Cifuentes-Muñoz N, et al. Virus Res. 2019 May;265:68-73. doi: 10.1016/j.virusres.2019.03.002. Epub 2019 Mar 5. Virus Res. 2019. PMID: 30844414 Free PMC article. Review. - [Human metapneumovirus as a common cause of respiratory tract disease].
Mazhul' LA, Schildgen O, Isaeva EI, Viazov SO. Mazhul' LA, et al. Vopr Virusol. 2007 May-Jun;52(3):4-8. Vopr Virusol. 2007. PMID: 17601042 Review. Russian.
Cited by
- Virus stealth technology: Tools to study virus cell-to-cell transmission.
Yin P, Martin CK, Kielian M. Yin P, et al. PLoS Pathog. 2024 Oct 9;20(10):e1012590. doi: 10.1371/journal.ppat.1012590. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39383183 Free PMC article. Review. No abstract available. - Tunneling Nanotubes: The Cables for Viral Spread and Beyond.
Kapoor D, Sharma P, Saini A, Azhar E, Elste J, Kohlmeir EK, Shukla D, Tiwari V. Kapoor D, et al. Results Probl Cell Differ. 2024;73:375-417. doi: 10.1007/978-3-031-62036-2_16. Results Probl Cell Differ. 2024. PMID: 39242387 Review. - Intercellular Highways in Transport Processes.
Szabó-Meleg E. Szabó-Meleg E. Results Probl Cell Differ. 2024;73:173-201. doi: 10.1007/978-3-031-62036-2_9. Results Probl Cell Differ. 2024. PMID: 39242380 Review. - Exploiting hosts and vectors: viral strategies for facilitating transmission.
Yu X, Zhu Y, Yin G, Wang Y, Shi X, Cheng G. Yu X, et al. EMBO Rep. 2024 Aug;25(8):3187-3201. doi: 10.1038/s44319-024-00214-6. Epub 2024 Jul 24. EMBO Rep. 2024. PMID: 39048750 Free PMC article. Review. - A role for tunneling nanotubes in virus spread.
Lv W, Li Z, Wang S, He J, Zhang L. Lv W, et al. Front Microbiol. 2024 Feb 16;15:1356415. doi: 10.3389/fmicb.2024.1356415. eCollection 2024. Front Microbiol. 2024. PMID: 38435698 Free PMC article. Review.
References
- Ebihara T, Endo R, Kikuta H, Ishiguro N, Yoshioka M, et al. (2003) Seroprevalence of human metapneumovirus in Japan. J Med Virol 70: 281–283. - PubMed
Grants and funding
- P20 GM103486/GM/NIGMS NIH HHS/United States
- P20 RR020171/RR/NCRR NIH HHS/United States
- R01 AI051517/AI/NIAID NIH HHS/United States
- R56 AI051517/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources