Upstream Freshwater and Terrestrial Sources Are Differentially Reflected in the Bacterial Community Structure along a Small Arctic River and Its Estuary - PubMed (original) (raw)
Upstream Freshwater and Terrestrial Sources Are Differentially Reflected in the Bacterial Community Structure along a Small Arctic River and Its Estuary
Aviaja L Hauptmann et al. Front Microbiol. 2016.
Abstract
Glacier melting and altered precipitation patterns influence Arctic freshwater and coastal ecosystems. Arctic rivers are central to Arctic water ecosystems by linking glacier meltwaters and precipitation with the ocean through transport of particulate matter and microorganisms. However, the impact of different water sources on the microbial communities in Arctic rivers and estuaries remains unknown. In this study we used 16S rRNA gene amplicon sequencing to assess a small river and its estuary on the Disko Island, West Greenland (69°N). Samples were taken in August when there is maximum precipitation and temperatures are high in the Disko Bay area. We describe the bacterial community through a river into the estuary, including communities originating in a glacier and a proglacial lake. Our results show that water from the glacier and lake transports distinct communities into the river in terms of diversity and community composition. Bacteria of terrestrial origin were among the dominating OTUs in the main river, while the glacier and lake supplied the river with water containing fewer terrestrial organisms. Also, more psychrophilic taxa were found in the community supplied by the lake. At the river mouth, the presence of dominant bacterial taxa from the lake and glacier was unnoticeable, but these taxa increased their abundances again further into the estuary. On average 23% of the estuary community consisted of indicator OTUs from different sites along the river. Environmental variables showed only weak correlations with community composition, suggesting that hydrology largely influences the observed patterns.
Keywords: Greenland; arctic; bacterial community; biodiversity; freshwater network; polar environments.
Figures
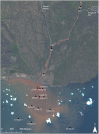
Figure 1
Sample sites, Disko Island, West Greenland, 69°N (Worldview, 2013).
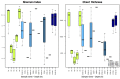
Figure 2
Shannon Index and Chao1 Richness. R1–R5 are river samples (n = 3), E100–E1100 are estuary samples from 100 to 1100 m into the estuary with varying number of replicates. Replicates are samples from the three different transects and should not be confused with replicates of the same water mass. Depth is indicated after each site name, such that for example E100-0.5 is taken at 0.5 m depth. Sample sites with no replicates (n = 1) are indicated without boxes, for sample sites with boxes n = 3.
Figure 3
NMDS plots of river data, estuary data and combined river and estuary data. Following stress values were obtained: river = 6.49%, estuary = 8.75%, combined = 10.29%. Depths are indicated at the estuary points.
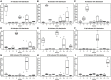
Figure 4
Percentage of indicator OTU sequences distribution across sample sites. Please note that Y-axes are different in each plot. (A–E) River sites (R1–R5) consist of three replicates corresponding to 36,540 sequences total for each site after rarefaction to 12,180 sequences per sample. (F–I) Estuary sample sites E300–E1100 are each sampled in three transects at two depths, making up six samples per distance from the river mouth, corresponding to 73,080 sequences. E100 lacks one sample at transect 1, deep sample, therefore it consists of 60,900 sequences.
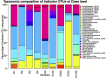
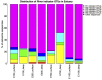
Figure 5
Taxonomic composition of indicator OTUs at Class level. Percentages of indicator OTU sequences of each Class are calculated as percentage of the total number of sequences of indicator OTUs from the individual samples.
Figure 6
Distribution of river indicator OTUs in the estuary. Calculated as percentage of river indicator OTU sequences of total estuary sequences of the estuary site in question. Note that the total number of sequences in each estuary site may differ due to different number of replicates as described in Section Materials and Methods.
Similar articles
- Terrestrial Inputs Shape Coastal Bacterial and Archaeal Communities in a High Arctic Fjord (Isfjorden, Svalbard).
Delpech LM, Vonnahme TR, McGovern M, Gradinger R, Præbel K, Poste AE. Delpech LM, et al. Front Microbiol. 2021 Feb 26;12:614634. doi: 10.3389/fmicb.2021.614634. eCollection 2021. Front Microbiol. 2021. PMID: 33717004 Free PMC article. - Spatial variation in bacterial community in natural wetland-river-sea ecosystems.
Zhang H, Zheng S, Ding J, Wang O, Liu F. Zhang H, et al. J Basic Microbiol. 2017 Jun;57(6):536-546. doi: 10.1002/jobm.201700041. Epub 2017 Apr 13. J Basic Microbiol. 2017. PMID: 28407285 - Highly Diverse Aquatic Microbial Communities Separated by Permafrost in Greenland Show Distinct Features According to Environmental Niches.
Bomberg M, Claesson Liljedahl L, Lamminmäki T, Kontula A. Bomberg M, et al. Front Microbiol. 2019 Jul 11;10:1583. doi: 10.3389/fmicb.2019.01583. eCollection 2019. Front Microbiol. 2019. PMID: 31354674 Free PMC article. - Strong Seasonality in Arctic Estuarine Microbial Food Webs.
Kellogg CTE, McClelland JW, Dunton KH, Crump BC. Kellogg CTE, et al. Front Microbiol. 2019 Nov 29;10:2628. doi: 10.3389/fmicb.2019.02628. eCollection 2019. Front Microbiol. 2019. PMID: 31849850 Free PMC article. Review. - Metagenomic insights into particles and their associated microbiota in a coastal margin ecosystem.
Simon HM, Smith MW, Herfort L. Simon HM, et al. Front Microbiol. 2014 Sep 5;5:466. doi: 10.3389/fmicb.2014.00466. eCollection 2014. Front Microbiol. 2014. PMID: 25250019 Free PMC article. Review.
Cited by
- Composition and Functional Characteristics and Influencing Factors of Bacterioplankton Community in the Huangshui River, China.
Zhang Q, Wu Z, Zhao J, Wang G, Hao J, Wang S, Lin Y, Guan H, Zhang J, Jian S, Li A. Zhang Q, et al. Microorganisms. 2021 Oct 29;9(11):2260. doi: 10.3390/microorganisms9112260. Microorganisms. 2021. PMID: 34835386 Free PMC article. - Contrasting Patterns of the Bacterial Communities in Melting Ponds and Periglacial Rivers of the Zhuxi glacier in the Tibet Plateau.
Hu Y, Yao X, Wu Y, Han W, Zhou Y, Tang X, Shao K, Gao G. Hu Y, et al. Microorganisms. 2020 Apr 2;8(4):509. doi: 10.3390/microorganisms8040509. Microorganisms. 2020. PMID: 32252494 Free PMC article. - Physicochemical Drivers of Microbial Community Structure in Sediments of Lake Hazen, Nunavut, Canada.
Ruuskanen MO, St Pierre KA, St Louis VL, Aris-Brosou S, Poulain AJ. Ruuskanen MO, et al. Front Microbiol. 2018 Jun 5;9:1138. doi: 10.3389/fmicb.2018.01138. eCollection 2018. Front Microbiol. 2018. PMID: 29922252 Free PMC article. - Microbial connectivity and sorting in a High Arctic watershed.
Comte J, Culley AI, Lovejoy C, Vincent WF. Comte J, et al. ISME J. 2018 Dec;12(12):2988-3000. doi: 10.1038/s41396-018-0236-4. Epub 2018 Aug 7. ISME J. 2018. PMID: 30087410 Free PMC article. - Integrated biogeography of planktonic and sedimentary bacterial communities in the Yangtze River.
Liu T, Zhang AN, Wang J, Liu S, Jiang X, Dang C, Ma T, Liu S, Chen Q, Xie S, Zhang T, Ni J. Liu T, et al. Microbiome. 2018 Jan 19;6(1):16. doi: 10.1186/s40168-017-0388-x. Microbiome. 2018. PMID: 29351813 Free PMC article.
References
- Bowman J. P., Nichols C. M., Gibson J. A. E. (2003). Algoriphagus ratkowskyi gen. nov., sp. nov., Brumimicrobium glaciale gen. nov., sp. nov., Cryomorpha ignava gen. nov., sp. nov. and Crocinitomix catalasitica gen. nov., sp. nov., novel flavobacteria isolated from various polar habitats. Int. J. Syst. Evol. Microbiol. 53, 1343–1355. 10.1099/ijs.0.02553-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources