SF2312 is a natural phosphonate inhibitor of enolase - PubMed (original) (raw)
. 2016 Dec;12(12):1053-1058.
doi: 10.1038/nchembio.2195. Epub 2016 Oct 10.
Nikunj Satani # 2, David Maxwell 3, Yu-Hsi Lin 2, Naima Hammoudi 2, Zhenghong Peng 4, Federica Pisaneschi 2, Todd M Link 1, Gilbert R Lee 4th 1, Duoli Sun 1, Basvoju A Bhanu Prasad 1, Maria Emilia Di Francesco 5, Barbara Czako 5, John M Asara 6, Y Alan Wang 7, William Bornmann 8, Ronald A DePinho 7, Florian L Muller 2
Affiliations
- PMID: 27723749
- PMCID: PMC5110371
- DOI: 10.1038/nchembio.2195
SF2312 is a natural phosphonate inhibitor of enolase
Paul G Leonard et al. Nat Chem Biol. 2016 Dec.
Abstract
Despite being crucial for energy generation in most forms of life, few if any microbial antibiotics specifically inhibit glycolysis. To develop a specific inhibitor of the glycolytic enzyme enolase 2 (ENO2) for the treatment of cancers with deletion of ENO1 (encoding enolase 1), we modeled the synthetic tool compound inhibitor phosphonoacetohydroxamate (PhAH) into the active site of human ENO2. A ring-stabilized analog of PhAH, in which the hydroxamic nitrogen is linked to Cα by an ethylene bridge, was predicted to increase binding affinity by stabilizing the inhibitor in a bound conformation. Unexpectedly, a structure-based search revealed that our hypothesized backbone-stabilized PhAH bears strong similarity to SF2312, a phosphonate antibiotic of unknown mode of action produced by the actinomycete Micromonospora, which is active under anaerobic conditions. Here, we present multiple lines of evidence, including a novel X-ray structure, that SF2312 is a highly potent, low-nanomolar inhibitor of enolase.
Figures
FIGURE 1. SF2312 is a potent inhibitor of Enolase with mixed competitive and non-competitive kinetics
(a) Chemical structure of PhAH, SF2312 (1), deoxy-SF2312 (2), with chiral centers indicated. (b) Enolase enzymatic activity was measured in lysates of the D423 cell line overexpressing human ENO1 and ENO2. Inhibitors were incubated with enzyme prior to addition of 5 mM substrate. Enzymatic activity was normalized to that in the absence of the inhibitor (y-axis, log2) and plotted as function of inhibitor concentration (nM, x-axis). Traces for ENO2 inhibited with SF2312, PhAH and deoxy-SF2312 are shown as red, blue and purple circles, respectively. Traces for ENO1 are shown crimson, light blue, and light purple diamonds. Each data point shows the mean ± S.D of N = 6 independent measurements. (c, d) Enolase activity (ENO2) was measured as a function of substrate (2-PGA) and inhibitor (c, SF2312; d, deoxy-SF2312) concentration. Data points show mean ± S.D of N = 5 independent measurements.
FIGURE 2. SF2312 stabilizes Enolase 2 against thermal denaturation
(a) Lysates of D423 overexpressing ENO2 cells were incubated with vehicle, 1 μM of PhAH or SF2312 and thermally denatured, with immunoblotting performed in post-centrifugation supernatants (uncropped images Supplementary Fig. 20). (b) Quantification of band intensity (y-axis) versus temperature (x-axis), with the trace in grey being vehicle, blue being PhAH and red being SF2312 treatment groups. (c) Mean melting temperatures (_T_m 50) ± S.D for N = 6 control, N = 4 PhAH, N = 6 SF2312 independent measurements. Significant results by unpaired t-test with Bonferroni correction are indicated.
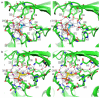
FIGURE 3. PhAH and SF2312 interact with ENO2 through a complex network of electrostatic, metal coordination and hydrogen bond interactions
(a) Stereo presentation of PhAH and (b) SF2312 binding in the ENO2 active site. The protein backbone is shown using the cartoon depiction with key amino acids for magnesium or ligand binding highlighted using the stick representation. Magnesium ions and water molecules are shown as magenta or red spheres respectively. Hydrogen bonds and metal coordination bonds are both represented by black dashed lines. The 2Fo-Fc unbiased omit electron density map for each ligand, cotoured at 1.5σ, is shown as a grey mesh around the ligand. Coordinates were deposited in PDB (ENO2:PhAH, 4ZA0; ENO2:SF2312, 4ZCW).
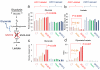
FIGURE 4. SF2312 inhibits Enolase activity and is selectively toxic to ENO1-deleted glioma cells
(a) Effect of SF2312 on cell proliferation (total cell number, Hoechst 33342) and (b) apoptosis (YO-PRO®-1 positive cells). D423 _ENO1_-deleted (red), D423 isogenic controls expressing ENO1 (dark blue) or overexpressing ENO2 (light blue) were treated for 2-weeks with varying concentrations of inhibitors as indicated (μM). Each condition was conducted in duplicate with each data point represents one biological replicate, expressed as a function of vehicle control. (c) A representative plate image stained with Hoechst 33342. The experiment probing toxicity of PhAH and SF2312 against D423 _ENO1_-deleted cells was repeated at least four times. (d) D423 _ENO1_-deleted and isogenic control glioma cells were treated with vehicle, 10 μM and 25 μM SF2312 for 72 hours. Polar metabolites were extracted and quantified by mass spec, showing the ratio of (d) phospho-creatine to creatine and (e) of 3-PGA to PEP. Bars represent individual measurements of independently treated cells. Light blue bars, D423 _ENO1_-deleted glioma cells CT; blue bars, _ENO1_-deleted cells treated with 10 μM SF2312; Dark blue bars, _ENO1_-deleted, 25 μM SF2312; Light crimson bars, D423 _ENO1_-rescued cells CT; crimson bars, _ENO1_-rescued, 10 μM SF2312; Brown bars, _ENO1_-rescued, 25 μM SF2312. Statistically significant comparisons, by unpaired, 2-tailed, t-test with Bonferroni correction are indicated.
FIGURE 5. SF2312 selectively blocks glycolysis in ENO1-deleted glioma cells
D423 _ENO1_-deleted and isogenic rescued cells were supplemented with 13C-1 labeled glucose and treated with 10 μM of SF2312 or PhAH for 72 hours. Integrals (expressed relative to the integral internal standard for NMR in D2O (3-(trimethylsilyl)propionic-2,2,3,3-d4) of peaks at 97 and 93 ppm (glucose, (a)), 20 ppm (C3 of lactate, (b)), 64 ppm (C3 of glycerate, (c)), and the ratio of glycerate to lactate (d) are shown (y-axis). Bars represent individual measurements of biological replicates. Significant differences by unpaired, two-tailed, t-test with Bonferroni correction are indicated. The experiment was conducted only once, but confirmed using 13C-uniformly labeled glucose (Supplementary Figure 9).
Similar articles
- The 3_S_ Enantiomer Drives Enolase Inhibitory Activity in SF2312 and Its Analogues.
Pisaneschi F, Lin YH, Leonard PG, Satani N, Yan VC, Hammoudi N, Raghavan S, Link TM, K Georgiou D, Czako B, Muller FL. Pisaneschi F, et al. Molecules. 2019 Jul 9;24(13):2510. doi: 10.3390/molecules24132510. Molecules. 2019. PMID: 31324042 Free PMC article. - Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Krucinska J, Lombardo MN, Erlandsen H, Hazeen A, Duay SS, Pattis JG, Robinson VL, May ER, Wright DL. Krucinska J, et al. Sci Rep. 2019 Nov 19;9(1):17106. doi: 10.1038/s41598-019-53301-3. Sci Rep. 2019. PMID: 31745118 Free PMC article. - Structure of the bis divalent cation complex with phosphonoacetohydroxamate at the active site of enolase.
Poyner RR, Reed GH. Poyner RR, et al. Biochemistry. 1992 Aug 11;31(31):7166-73. doi: 10.1021/bi00146a020. Biochemistry. 1992. PMID: 1322695 - Beyond ENO1, emerging roles and targeting strategies of other enolases in cancers.
Ni J, Huang Y, Li C, Yin Q, Ying J. Ni J, et al. Mol Ther Oncolytics. 2023 Nov 10;31:100750. doi: 10.1016/j.omto.2023.100750. eCollection 2023 Dec 19. Mol Ther Oncolytics. 2023. PMID: 38075246 Free PMC article. Review. - The behavior and significance of slow-binding enzyme inhibitors.
Morrison JF, Walsh CT. Morrison JF, et al. Adv Enzymol Relat Areas Mol Biol. 1988;61:201-301. doi: 10.1002/9780470123072.ch5. Adv Enzymol Relat Areas Mol Biol. 1988. PMID: 3281418 Review. No abstract available.
Cited by
- ZFP64 drives glycolysis-mediated stem cell-like properties and tumorigenesis in breast cancer.
Sun J, Liu J, Hou Y, Bao J, Wang T, Liu L, Zhang Y, Zhong R, Sun Z, Ye Y, Liu J. Sun J, et al. Biol Direct. 2024 Sep 18;19(1):83. doi: 10.1186/s13062-024-00533-7. Biol Direct. 2024. PMID: 39294751 Free PMC article. - Deciphering T Cell Immunometabolism with Activity-Based Protein Profiling.
Borne AL, Huang T, McCloud RL, Pachaiyappan B, Bullock TNJ, Hsu KL. Borne AL, et al. Curr Top Microbiol Immunol. 2019;420:175-210. doi: 10.1007/82_2018_124. Curr Top Microbiol Immunol. 2019. PMID: 30128827 Free PMC article. Review. - In silico prediction of a new lead compound targeting enolase of trypanosomatids through structure-based virtual screening and molecular dynamic studies.
Vidhya VM, Lakshmi BS, Ponnuraj K. Vidhya VM, et al. J Mol Model. 2020 Jan 7;26(2):23. doi: 10.1007/s00894-019-4284-0. J Mol Model. 2020. PMID: 31912304 - Glycometabolic rearrangements--aerobic glycolysis in pancreatic cancer: causes, characteristics and clinical applications.
Cao L, Wu J, Qu X, Sheng J, Cui M, Liu S, Huang X, Xiang Y, Li B, Zhang X, Cui R. Cao L, et al. J Exp Clin Cancer Res. 2020 Nov 30;39(1):267. doi: 10.1186/s13046-020-01765-x. J Exp Clin Cancer Res. 2020. PMID: 33256814 Free PMC article. Review. - Metabolic collateral lethal target identification reveals MTHFD2 paralogue dependency in ovarian cancer.
Achreja A, Yu T, Mittal A, Choppara S, Animasahun O, Nenwani M, Wuchu F, Meurs N, Mohan A, Jeon JH, Sarangi I, Jayaraman A, Owen S, Kulkarni R, Cusato M, Weinberg F, Kweon HK, Subramanian C, Wicha MS, Merajver SD, Nagrath S, Cho KR, DiFeo A, Lu X, Nagrath D. Achreja A, et al. Nat Metab. 2022 Sep;4(9):1119-1137. doi: 10.1038/s42255-022-00636-3. Epub 2022 Sep 21. Nat Metab. 2022. PMID: 36131208
References
- Fothergill-Gilmore LA, Michels PA. Evolution of glycolysis. Progress in biophysics and molecular biology. 1993;59:105–235. - PubMed
ONLINE METHODS REFERENCES
- Brewer JM. Yeast enolase: mechanism of activation by metal ions. CRC Crit. Rev. Biochem. 1981;11:209–254. - PubMed
- Schrödinger Suite 2013-3 Virtual Screening Workflow. Glide version 5.9, LigPrep version 2.6. Schrödinger, LLC; New York, NY: 2013. (Schrödinger, LLC, New York, NY, 2013.)
- Prime. Version 3.4 v. 3.4 Schrödinger, LLC; New York, NY: 2013.
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- P01 CA095616/CA/NCI NIH HHS/United States
- P01 CA120964/CA/NCI NIH HHS/United States
- P50 CA127001/CA/NCI NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous