Comprehensive Cross-Population Analysis of High-Grade Serous Ovarian Cancer Supports No More Than Three Subtypes - PubMed (original) (raw)
Comprehensive Cross-Population Analysis of High-Grade Serous Ovarian Cancer Supports No More Than Three Subtypes
Gregory P Way et al. G3 (Bethesda). 2016.
Abstract
Four gene expression subtypes of high-grade serous ovarian cancer (HGSC) have been previously described. In these early studies, a fraction of samples that did not fit well into the four subtype classifications were excluded. Therefore, we sought to systematically determine the concordance of transcriptomic HGSC subtypes across populations without removing any samples. We created a bioinformatics pipeline to independently cluster the five largest mRNA expression datasets using k-means and nonnegative matrix factorization (NMF). We summarized differential expression patterns to compare clusters across studies. While previous studies reported four subtypes, our cross-population comparison does not support four. Because these results contrast with previous reports, we attempted to reproduce analyses performed in those studies. Our results suggest that early results favoring four subtypes may have been driven by the inclusion of serous borderline tumors. In summary, our analysis suggests that either two or three, but not four, gene expression subtypes are most consistent across datasets.
Keywords: molecular subtypes; ovarian cancer; reproducibility; unsupervised clustering.
Copyright © 2016 Way et al.
Figures
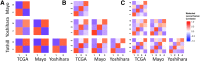
Figure 1
Significance analysis of microarray (SAM) moderated t score Pearson correlation heatmaps reveal consistency across datasets. (A) Correlations across datasets for k means k = 2. (B) Correlations across datasets for k means k = 3. (C) Correlations across datasets for k means k = 4. TCGA, The Cancer Genome Atlas.
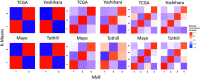
Figure 2
Significance analysis of microarray (SAM) moderated t score Pearson correlation heatmaps of clusters formed by k means clustering and NMF clustering reveals consistency across clustering methods. Within dataset results are shown for both methods when setting each algorithm to find 2, 3, and 4 clusters. NMF, nonnegative matrix factorization; TCGA, The Cancer Genome Atlas.
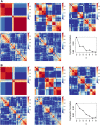
Figure 3
Comparing NMF consensus clustering in the Tothill dataset. Data displays consensus clustering for k = 2 to k = 6 for 10 NMF initializations alongside the cophenetic correlation results for k = 2 to k = 8. (A) Tothill dataset (n = 260) with borderline samples (n = 18) not removed prior to clustering. (B) Tothill dataset with borderline samples removed (n = 242).
Similar articles
- Combination of TP53 and AGR3 to distinguish ovarian high-grade serous carcinoma from low-grade serous carcinoma.
Qiu C, Wang Y, Wang X, Zhang Q, Li Y, Xu Y, Jin C, Bu H, Zheng W, Yang X, Lu N, Kong B. Qiu C, et al. Int J Oncol. 2018 Jun;52(6):2041-2050. doi: 10.3892/ijo.2018.4360. Epub 2018 Apr 4. Int J Oncol. 2018. PMID: 29620196 - Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53-mutant tumors and identifies NRAS as an oncogenic driver.
Emmanuel C, Chiew YE, George J, Etemadmoghadam D, Anglesio MS, Sharma R, Russell P, Kennedy C, Fereday S, Hung J, Galletta L, Hogg R, Wain GV, Brand A, Balleine R, MacConaill L, Palescandolo E, Hunter SM, Campbell I, Dobrovic A, Wong SQ, Do H, Clarke CL, Harnett PR, Bowtell DD, deFazio A; Australian Ovarian Cancer Study (AOCS). Emmanuel C, et al. Clin Cancer Res. 2014 Dec 15;20(24):6618-30. doi: 10.1158/1078-0432.CCR-14-1292. Epub 2014 Oct 14. Clin Cancer Res. 2014. PMID: 25316818 - Gene co-expression network reveals shared modules predictive of stage and grade in serous ovarian cancers.
Sun Q, Zhao H, Zhang C, Hu T, Wu J, Lin X, Luo D, Wang C, Meng L, Xi L, Li K, Hu J, Ma D, Zhu T. Sun Q, et al. Oncotarget. 2017 Jun 27;8(26):42983-42996. doi: 10.18632/oncotarget.17785. Oncotarget. 2017. PMID: 28562334 Free PMC article. - Low-grade and high-grade serous Mullerian carcinoma: review and analysis of publicly available gene expression profiles.
May T, Shoni M, Crum CP, Xian W, Vathipadiekal V, Birrer M, Rosen B, Tone A, Murphy KJ. May T, et al. Gynecol Oncol. 2013 Mar;128(3):488-92. doi: 10.1016/j.ygyno.2012.12.009. Epub 2012 Dec 16. Gynecol Oncol. 2013. PMID: 23253401 Review. - Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis.
McCluggage WG. McCluggage WG. Pathology. 2011 Aug;43(5):420-32. doi: 10.1097/PAT.0b013e328348a6e7. Pathology. 2011. PMID: 21716157 Review.
Cited by
- Subject level clustering using a negative binomial model for small transcriptomic studies.
Li Q, Noel-MacDonnell JR, Koestler DC, Goode EL, Fridley BL. Li Q, et al. BMC Bioinformatics. 2018 Dec 12;19(1):474. doi: 10.1186/s12859-018-2556-9. BMC Bioinformatics. 2018. PMID: 30541426 Free PMC article. - Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma.
Chen GM, Kannan L, Geistlinger L, Kofia V, Safikhani Z, Gendoo DMA, Parmigiani G, Birrer M, Haibe-Kains B, Waldron L. Chen GM, et al. Clin Cancer Res. 2018 Oct 15;24(20):5037-5047. doi: 10.1158/1078-0432.CCR-18-0784. Epub 2018 Jul 3. Clin Cancer Res. 2018. PMID: 30084834 Free PMC article. - SGK1 suppresses ferroptosis in ovarian cancer via NRF2-dependent and -independent pathways.
Sang X, Han J, Wang Z, Cai W, Liao X, Kong Z, Yu Z, Cheng H, Liu P. Sang X, et al. Oncogene. 2024 Nov;43(45):3335-3347. doi: 10.1038/s41388-024-03173-3. Epub 2024 Sep 21. Oncogene. 2024. PMID: 39306614 - Machine learning-based integration develops an immune-related risk model for predicting prognosis of high-grade serous ovarian cancer and providing therapeutic strategies.
Wu Q, Tian R, He X, Liu J, Ou C, Li Y, Fu X. Wu Q, et al. Front Immunol. 2023 Apr 5;14:1164408. doi: 10.3389/fimmu.2023.1164408. eCollection 2023. Front Immunol. 2023. PMID: 37090728 Free PMC article. - Functional network community detection can disaggregate and filter multiple underlying pathways in enrichment analyses.
Harrington LX, Way GP, Doherty JA, Greene CS. Harrington LX, et al. Pac Symp Biocomput. 2018;23:157-167. Pac Symp Biocomput. 2018. PMID: 29218878 Free PMC article.
References
- Boettiger C., 2015. An introduction to Docker for reproducible research. ACM SIGOPS Oper. Syst. Rev. 49: 71–79.
- Bonome T., Lee J.-Y., Park D.-C., Radonovich M., Pise-Masison C., et al. , 2005. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 65: 10602–10612. - PubMed
- Broad Institute TCGA Genome Data Analysis Center, 2016a Analysis overview for ovarian serous cystadenocarcinoma (primary solid tumor cohort) - 28 January 2016. Broad Institute of MIT and Harvard. DOI: 10.7908/C1VQ324T.
Publication types
MeSH terms
Grants and funding
- F31 CA186625/CA/NCI NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- R01 CA122443/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- R01 CA200854/CA/NCI NIH HHS/United States
- R01 CA168758/CA/NCI NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases