A Minimum Epitope Overlap between Infections Strongly Narrows the Emerging T Cell Repertoire - PubMed (original) (raw)
A Minimum Epitope Overlap between Infections Strongly Narrows the Emerging T Cell Repertoire
Susanne G Oberle et al. Cell Rep. 2016.
Abstract
Many infections are caused by pathogens that are similar, but not identical, to previously encountered viruses, bacteria, or vaccines. In such re-infections, pathogens introduce known antigens, which are recognized by memory T cells and new antigens that activate naive T cells. How preexisting memory T cells impact the repertoire of T cells responding to new antigens is still largely unknown. We demonstrate that even a minimum epitope overlap between infections strongly increases the activation threshold and narrows the diversity of T cells recruited in response to new antigens. Thus, minimal cross-reactivity between infections can significantly impact the outcome of a subsequent immune response. Interestingly, we found that non-transferrable memory T cells are most effective in raising the activation threshold. Our findings have implications for designing vaccines and suggest that vaccines meant to target low-affinity T cells are less effective when they contain a strong CD8 T cell epitope that has previously been encountered.
Keywords: T cell activation threshold; cytotoxic T cells; secondary immune responses; strength of TCR stimulation.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
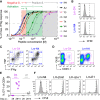
Figure 1
Only High-Affinity T Cells Expand in _Listeria_-Immune Mice (A) Schematic illustration of the Experimental Procedures. Naive controls and C57BL/6 mice infected with wild-type Listeria (Lm-WT) received 104 naive OT-1 4 weeks later and the indicated Listeria strains. (B) Frequency of OT-1 T cells among peripheral blood CD8+ T cells 6 days after infection. Numbers indicate the ratio of OT-1 T cells in naive and Lm-WT immune hosts. (C) Naive or Lm-WT immune mice grafted with 2.5 × 105 CFSE-labeled OT-1 cells and infected with Lm-N4 or Lm-T4. CFSE dilution among splenic OT-1 (filled histogram) was measured 78 hr later. Dashed lines are unlabeled host CD8+ T cells. Data are representative of three (B) or two (C) independent experiments with three to five mice per group. (D) Naive or Lm-WT infected Rip-mOva mice were 4 weeks later engrafted with low-affinity H-2Kb/Ova-specific OT-3 TCR transgenic T cells and then infected with Lm-N4. Blood glucose levels were determined 8 days later. Statistical analysis are unpaired Student’s t tests with ∗∗p < 0.01 and ∗∗∗∗p < 0.0001.
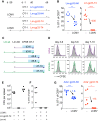
Figure 2
Even Extremely Low-Affinity Stimulation Induces Normal Activation and Memory Generation (A) Dose response curves graphing peptide concentration against the fraction of maximum numbers of IFNγ producing T cells were obtained by briefly stimulating an OT-1 T cell line in vitro with the indicated peptides. Note that only a fraction of the OT-1 T cells produced IFNγ after very low-affinity ligand stimulation. Data were therefore normalized to better compare the EC50. (B) C57BL/6 mice transferred with 2 × 105 CFSE-labeled OT-1 cells and infected the indicated Listeria strains were analyzed for splenic CFSE dilution at 76 hr post infection. (C and D) Naive mice received 105 OT-1 T cells and an Lm-D4 or Lm-N4 infection. Splenocytes were 4.5 days later and briefly re-stimulated with SIINFEKL peptide and stained intracellularly for IFNγ, TNF (C), and GrzB (D). (C) shows representative OT-1 and (D) CD8+ gated flow cytometry plots. (E) Naive mice received 2 × 104 OT-1 and the indicated Listeria strains and OT-1 frequency among peripheral blood CD8+ T cells was determined 28 days later. The mice were then infected with VSV-N4, and the OT-1 frequency was measured 4 days later in the blood. (F) CFSE+ OT-1-containing mice were infected with Lm-N4, Lm-Cpα1, Lm-βcat, or Lm-E1 and analyzed as described in (B). The data are representative for four (A) or three independent experiments with three to five mice per group (B, C, and D) or two experiments with two to five mice per group (E and F). Statistical analysis are unpaired Student’s t tests, ∗∗∗∗p < 0.0001.
Figure 3
Memory CD8+ T Cells Raise the Activation Threshold in Secondary Infections Independently of the Inflammatory Milieu (A and B) Control (–) or LCMV-immune mice received 4 weeks after the infection 104 OT-1 and Listeria co-expressing the LCMV gp33 peptide and N4 Ova (Lm-gp33-N4) or gp33 and T4 Ova (Lm-gp33-T4). Note that only the gp33 and gp34 epitopes are shared between the consecutive infections. Frequency of blood OT-1 T cells among total CD8+ T cells was measured 6 days later. Numbers indicate the OT-1 frequency fold change between naive and immune hosts. (C and D) 3 × 105 naive CFSE-labeled OT-1 cells were transferred at different time points into ongoing primary Lm-N4 or secondary Lm-WT and Lm-N4 infections in order to assess antigen presentation kinetics. CFSE dilution was measured 80 hr after the transfer. (E and F) Naive and LCMV-immune mice were infected with 2,000 or 2 × 105 CFU Lm-gp33-T4. Another LCMV-immune group was co-infected with 2 × 105 CFU Lm-gp33-T4 plus 2,000 CFU Lm-WT. Splenic Listeria load was determined 4 days later (E), and the OT-1 frequency among total blood CD8+ T cells was determined 6 days later (F). (G) Naive (–) or LCMV-immune (LCMV) mice received 104 OT-1 and 106 CFU _ActA_-deficient Listeria (_ActA_−gp33-N4 or _ActA_−gp33-T4). The OT-1 frequency among total blood CD8+ T cells was determined 6 days later. Numbers indicate the OT-1 ratio in naive and immune hosts. The data are representative for two (A–F) or one (G) independent experiments with n = 3–5 per group. Statistical analysis are unpaired Student’s t tests with ∗p < 0.1, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
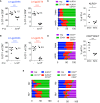
Figure 4
Endogenous Memory CD8+ T Cells Effectively Raise the Activation Threshold (A and B) All mice received 104 OT-1 T cells and some 3 × 105 (A) or 106 (B) memory P14 T cells. The OT-1 frequency among total CD8+ T cells was determined 7 days after infecting the mice with Lm-gp33-N4 or Lm-gp33-T4. Data are representative for two independent experiments with n = 5 mice per group. (C–E) CD45.1 P14 memory T cells were obtained from mice infected 30–90 days earlier with LCMV and transferred into naive mice. Shown are memory P14 T cell populations before and after the transfer. Bars represent individual mice, and the relative distribution of the indicated P14 subsets. The plots on the right show the frequency of KLRG1+ (C) and CD27+CD43+ subpopulations (D). Dots represent individual mice. (E) Mice with endogenous memory cells were i.v. injected with fluorescently labeled anti-CD8 antibody and sacrificed 3 min. later. CD127/KLRG1 and CD27/CD43 expression status on antibody-positive and -negative cells are shown as indicated in (D). Data are representative for two to four independent experiments with n = 3–5 mice per group. Statistical analyses are unpaired Student’s t tests with ∗p < 0.1, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Similar articles
- Epitope-specific regulation of memory programming by differential duration of antigen presentation to influenza-specific CD8(+) T cells.
Ballesteros-Tato A, León B, Lee BO, Lund FE, Randall TD. Ballesteros-Tato A, et al. Immunity. 2014 Jul 17;41(1):127-40. doi: 10.1016/j.immuni.2014.06.007. Immunity. 2014. PMID: 25035957 Free PMC article. - Inefficient cross-presentation limits the CD8+ T cell response to a subdominant tumor antigen epitope.
Otahal P, Hutchinson SC, Mylin LM, Tevethia MJ, Tevethia SS, Schell TD. Otahal P, et al. J Immunol. 2005 Jul 15;175(2):700-12. doi: 10.4049/jimmunol.175.2.700. J Immunol. 2005. PMID: 16002665 - Direct visualization of cross-reactive effector and memory allo-specific CD8 T cells generated in response to viral infections.
Brehm MA, Markees TG, Daniels KA, Greiner DL, Rossini AA, Welsh RM. Brehm MA, et al. J Immunol. 2003 Apr 15;170(8):4077-86. doi: 10.4049/jimmunol.170.8.4077. J Immunol. 2003. PMID: 12682237 - Lack of original antigenic sin in recall CD8(+) T cell responses.
Zehn D, Turner MJ, Lefrançois L, Bevan MJ. Zehn D, et al. J Immunol. 2010 Jun 1;184(11):6320-6. doi: 10.4049/jimmunol.1000149. Epub 2010 May 3. J Immunol. 2010. PMID: 20439913 Free PMC article.
Cited by
- Convergent clonal selection of donor- and recipient-derived CMV-specific T cells in hematopoietic stem cell transplant patients.
Erickson JR, Stevens-Ayers T, Mair F, Edmison B, Boeckh M, Bradley P, Prlic M. Erickson JR, et al. Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2117031119. doi: 10.1073/pnas.2117031119. Proc Natl Acad Sci U S A. 2022. PMID: 35105810 Free PMC article. - Expression of the DNA-Binding Factor TOX Promotes the Encephalitogenic Potential of Microbe-Induced Autoreactive CD8+ T Cells.
Page N, Klimek B, De Roo M, Steinbach K, Soldati H, Lemeille S, Wagner I, Kreutzfeldt M, Di Liberto G, Vincenti I, Lingner T, Salinas G, Brück W, Simons M, Murr R, Kaye J, Zehn D, Pinschewer DD, Merkler D. Page N, et al. Immunity. 2018 May 15;48(5):937-950.e8. doi: 10.1016/j.immuni.2018.04.005. Immunity. 2018. PMID: 29768177 Free PMC article. - The Role of Pre-existing Cross-Reactive Central Memory CD4 T-Cells in Vaccination With Previously Unseen Influenza Strains.
Nienen M, Stervbo U, Mölder F, Kaliszczyk S, Kuchenbecker L, Gayova L, Schweiger B, Jürchott K, Hecht J, Neumann AU, Rahmann S, Westhoff T, Reinke P, Thiel A, Babel N. Nienen M, et al. Front Immunol. 2019 Apr 4;10:593. doi: 10.3389/fimmu.2019.00593. eCollection 2019. Front Immunol. 2019. PMID: 31019503 Free PMC article. - DNA probes that store mechanical information reveal transient piconewton forces applied by T cells.
Ma R, Kellner AV, Ma VP, Su H, Deal BR, Brockman JM, Salaita K. Ma R, et al. Proc Natl Acad Sci U S A. 2019 Aug 20;116(34):16949-16954. doi: 10.1073/pnas.1904034116. Epub 2019 Aug 7. Proc Natl Acad Sci U S A. 2019. PMID: 31391300 Free PMC article. - Strong homeostatic TCR signals induce formation of self-tolerant virtual memory CD8 T cells.
Drobek A, Moudra A, Mueller D, Huranova M, Horkova V, Pribikova M, Ivanek R, Oberle S, Zehn D, McCoy KD, Draber P, Stepanek O. Drobek A, et al. EMBO J. 2018 Jul 13;37(14):e98518. doi: 10.15252/embj.201798518. Epub 2018 May 11. EMBO J. 2018. PMID: 29752423 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials