Molecular Evolution of Human Coronavirus Genomes - PubMed (original) (raw)
Review
Molecular Evolution of Human Coronavirus Genomes
Diego Forni et al. Trends Microbiol. 2017 Jan.
Abstract
Human coronaviruses (HCoVs), including SARS-CoV and MERS-CoV, are zoonotic pathogens that originated in wild animals. HCoVs have large genomes that encode a fixed array of structural and nonstructural components, as well as a variety of accessory proteins that differ in number and sequence even among closely related CoVs. Thus, in addition to recombination and mutation, HCoV genomes evolve through gene gains and losses. In this review we summarize recent findings on the molecular evolution of HCoV genomes, with special attention to recombination and adaptive events that generated new viral species and contributed to host shifts and to HCoV emergence. VIDEO ABSTRACT.
Keywords: gene gain/loss; host shift; human coronavirus; molecular evolution; positive selection; recombination.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
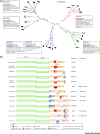
Figure 1
Key Figure: Phylogenetic Relationships and Genome Organization of Human and Animal Coronaviruses (CoVs) CoVs that infect nonhuman mammals are included only if they are mentioned in the text for comparative purposes. (A) The phylogenetic tree of complete genome sequences of HCoVs and selected mammalian CoVs was obtained with RAxML 8.2.4 . Numbers indicate bootstrap support. CoVs are colored according to genus and lineage. Information about origin, intermediate host, and clinical presentation is reported for the six HCoVs , , , , , . Data about case fatality rate were derived from the World Health Organization website (
). (B) CoV genome organization is schematically reported together with information on receptor/coreceptor usage. Virus names are colored according to their genus or lineage, as in (A). Only ORFs mentioned in the text are colored or shaded. Empty boxes represent accessory ORFs that are not described in the text.
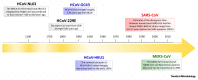
Figure I
Timeline for the Emergence of HCoVs.
Figure 2
Evolution of Human Coronavirus (HCoV) Accessory Proteins.(A) Test for relaxation of selective strength for SARS-CoV and SARSr-BatCoVs ORF8. Branches are colored according to the selection intensity parameter k. RELAX evaluates if selection on the test branches (bold) is relaxed (k < 1) or intensified (_k_ > 1) compared to background branches. In the evolutionary analysis table the number of sequences differs from that in the tree because RELAX removes identical sequences. Evidence of positive selection was searched for using the M7/M8 ‘site models’ from PAML (see Box 2) and with PARRIS. M7 and M8 represent the null and the positive selection models, respectively. A likelihood ratio test (with 2 degrees of freedom) was applied. (B) An amino acid alignment of rodent AKAP7 and four viral phosphodiesterases (PDEs) is shown. Amino acids are colored red if they are identical, orange if they have very similar properties. PDEs belonging to the 2H family are characterized by two H-Φ-[S/T]-Φ motifs (blue boxes), where Φ is a hydrophobic residue. The structure of rat AKAP7 (gray, PDB ID: 2VFK) is superimposed on MERS-CoV NS4b (green, model generated from 2VFK), MHV NS2a (cyan, PDB ID: 4Z5V), and Rotavirus A VP3 (yellow, PDB ID: 5AF2). Catalytic histidines are shown in red. (C) Sequence and membrane topology comparison of HCoV viroporins. Transmembrane regions (TM1-3) predicted by the TMHMM algorithm are boxed in blue. The corresponding topology model for SARS-CoV ORF3A, HCoV-229E ORF4a (from the Inf-1 strain), and HCoV-NL63 ORF3 is shown. The topology model of HCoV-OC43 OFR5 was derived from recent data .
Figure 3
Evolution at the Coronavirus (CoV)–Host Interaction Surface.(A) Schematic representation of MERS-CoV spike protein domains. Positively selected sites in MERS-CoV and other lineage C beta-CoVs are shown in red, RBD mutations emerged in the South Korean outbreak are in magenta (see text). A detail of the interaction surface between the MERS-CoV RBD and human DPP4 (PDB ID: 4F5C) is also reported. (B) Ribbon diagram of the interaction surface of human ACE2 with the spike protein of SARS-CoV (PDB ID: 2AJF) and HCoV-NL63 (PDB ID: 3KBH). The binding surface of porcine ANPEP with the TGEV spike protein (PDB ID: 4F5C) is also shown. The location of the HCoV-299E binding site on ANPEP is circled. Red denotes protein regions involved in binding.
Similar articles
- A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic.
Pollett S, Conte MA, Sanborn M, Jarman RG, Lidl GM, Modjarrad K, Maljkovic Berry I. Pollett S, et al. Sci Rep. 2021 Aug 30;11(1):17365. doi: 10.1038/s41598-021-96626-8. Sci Rep. 2021. PMID: 34462471 Free PMC article. - Zoonotic origins of human coronaviruses.
Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Ye ZW, et al. Int J Biol Sci. 2020 Mar 15;16(10):1686-1697. doi: 10.7150/ijbs.45472. eCollection 2020. Int J Biol Sci. 2020. PMID: 32226286 Free PMC article. Review. - Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination.
Lau SK, Feng Y, Chen H, Luk HK, Yang WH, Li KS, Zhang YZ, Huang Y, Song ZZ, Chow WN, Fan RY, Ahmed SS, Yeung HC, Lam CS, Cai JP, Wong SS, Chan JF, Yuen KY, Zhang HL, Woo PC. Lau SK, et al. J Virol. 2015 Oct;89(20):10532-47. doi: 10.1128/JVI.01048-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269185 Free PMC article. - Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses.
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Su S, et al. Trends Microbiol. 2016 Jun;24(6):490-502. doi: 10.1016/j.tim.2016.03.003. Epub 2016 Mar 21. Trends Microbiol. 2016. PMID: 27012512 Free PMC article. Review.
Cited by
- A review: Epidemiology, pathogenesis and prospect in developing vaccines for novel Coronavirus (COVID-19).
Gupta P. Gupta P. Indian J Tuberc. 2021 Jan;68(1):92-98. doi: 10.1016/j.ijtb.2020.09.021. Epub 2020 Sep 28. Indian J Tuberc. 2021. PMID: 33641858 Free PMC article. Review. - Review of the risk factors for SARS-CoV-2 transmission.
Li X, Xia WY, Jiang F, Liu DY, Lei SQ, Xia ZY, Wu QP. Li X, et al. World J Clin Cases. 2021 Mar 6;9(7):1499-1512. doi: 10.12998/wjcc.v9.i7.1499. World J Clin Cases. 2021. PMID: 33728294 Free PMC article. Review. - Acetylated K676 TGFBIp as a severity diagnostic blood biomarker for SARS-CoV-2 pneumonia.
Park HH, Kim HN, Kim H, Yoo Y, Shin H, Choi EY, Bae JS, Lee W. Park HH, et al. Sci Adv. 2020 Jul 31;6(31):eabc1564. doi: 10.1126/sciadv.abc1564. Print 2020 Jul. Sci Adv. 2020. PMID: 32937590 Free PMC article. - Structural and Drug Screening Analysis of the Non-structural Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 Virus Extracted From Indian Coronavirus Disease 2019 Patients.
Biswas N, Kumar K, Mallick P, Das S, Kamal IM, Bose S, Choudhury A, Chakrabarti S. Biswas N, et al. Front Genet. 2021 Mar 9;12:626642. doi: 10.3389/fgene.2021.626642. eCollection 2021. Front Genet. 2021. PMID: 33767730 Free PMC article. - Genome based evolutionary lineage of SARS-CoV-2 towards the development of novel chimeric vaccine.
Akhand MRN, Azim KF, Hoque SF, Moli MA, Joy BD, Akter H, Afif IK, Ahmed N, Hasan M. Akhand MRN, et al. Infect Genet Evol. 2020 Nov;85:104517. doi: 10.1016/j.meegid.2020.104517. Epub 2020 Sep 1. Infect Genet Evol. 2020. PMID: 32882432 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous