Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism - PubMed (original) (raw)
Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism
Jung-Eun Park et al. Proc Natl Acad Sci U S A. 2016.
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) infects humans from zoonotic sources and causes severe pulmonary disease. Virions require spike (S) glycoproteins for binding to cell receptors and for catalyzing virus-cell membrane fusion. Fusion occurs only after S proteins are cleaved sequentially, first during their secretion through the exocytic organelles of virus-producing cells, and second after virus binding to target-cell receptors. To more precisely determine how sequential proteolysis contributes to CoV infection, we introduced S mutations obstructing the first cleavages. These mutations severely compromised MERS-CoV infection into human lung-derived cells, but had little effect on infection into several other cell types. These cell type-specific requirements for proteolysis correlated with S conformations during cell entry. Without the first cleavages, S proteins resisted cell receptor-induced conformational changes, which restricted the second, fusion-activating cleavages. Consistent with these findings, precleaved MERS viruses used receptor-proximal, cell-surface proteases to effect the second fusion-activating cleavages during cell entry, whereas the more rigid uncleaved MERS viruses trafficked past these cell-surface proteases and into endosomes. Uncleaved viruses were less infectious to human airway epithelial and Calu3 cell cultures because they lacked sufficient endosomal fusion-activating proteases. Thus, by sensitizing viruses to receptor-induced conformational changes, the first S cleavages expand virus tropism to cell types that are relevant to lung infection, and therefore may be significant determinants of MERS-CoV virulence.
Keywords: coronavirus; protease; receptor; virus entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
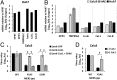
Fig. 1.
S1/S2 cleavage is required for MERS-CoV infection of Calu3 cells. (A) CoV particles have ∼100 S trimers. A single CoV S monomer is highlighted. A globular RBD (S1) and a stem-like FD (S2) is separated by domains that include the S1/S2 cleavage site. (B) A linear depiction of the MERS-CoV S protein includes a RBD, FP, two heptad repeats (HR1 and HR2), and a transmembrane domain (TMD). Two known proteolytic cleavage sites (S1/S2 and S2′) are depicted. Additional putative furin/proprotein convertase (black arrow heads) and cathepsin (white arrow heads) cleavage sites were predicted by ProP 1.0 Server (
) and SitePrediction (
www.dmbr.ugent.be/prx/bioit2-public/SitePrediction/index.php
), respectively. (C) Vero81, Huh7, and Calu3 cells were infected with WT and S1/S2 mutant (YSAS and SSVR) MERS-CoVs. After 5 h, subgenomic N mRNAs were determined by real-time PCR, and plotted relative to N mRNA levels in WT MERS-CoV–infected Vero81 cells. (D) Vero81, Huh7, and Calu3 cells were infected with WT and S1/S2 mutant MERS-CoVs. Progeny were collected at 20-h postinfection and titrated by plaque assay in Vero81 cells. (E) Six HAE cultures were infected with WT and S1/S2 mutant MERS-CoVs. Progeny were collected at 20-h postinfection and titrated by plaque assay in Vero81 cells. Error bars present SD from the mean (n = 3 for cultured cells, n = 6 for HAE cultures). Statistical significance was assessed by Student's t test. †P < 0.01; ‡P < 0.001; ns, not significant. (F) Vero81, Huh7, and Calu3 cells were infected with WT and S1/S2 mutant MERS-CoVs and S proteins within cells were analyzed by Western blot. Uncleaved (Sunc) and cleaved (S1 and S2) positions are indicated. The numbers at the left indicate molecular mass in kilodaltons.
Fig. 2.
S1/S2 cleavage is required for MERS pp entry into Calu3 cells. (A) Vero81, Huh7, and Calu3 cells were inoculated with WT, S1/S2 mutant (YSAS and SSVR) MERS pps, or with pp-lacking S proteins (Bald). (B) Three HAE cultures were inoculated with VSV-based WT and S1/S2 mutant MERS pseuodoparticles. (C) MERS pps were pretreated with trypsin and then used to inoculate Calu3 cells. (D) MERS pps were produced in the presence of PCI, cleared free of residual PCI, and used to inoculate Calu3 cells. In all experiments, virus entry was quantified by measuring luciferase levels at 18 h (B) or at 48 h (A, C, D) posttransduction. Lower panels depict S proteins on MERS pps after Western blotting. Uncleaved (Sunc) and cleaved (S2) positions are indicated. The numbers at the left indicate molecular mass in kilodaltons. Error bars present SD from the mean (n = 3). Statistical significance was assessed by Student's t test. *P < 0.05; †P < 0.01; ‡P < 0.001; ns, not significant.
Fig. 3.
SARS-CoV S cleavage by trypsin facilitates entry into Calu3 cells. (A) SARS and HCoV-229E pps were treated with trypsin. S proteins on pps were analyzed by Western blot. Uncleaved (Sunc) and cleaved (S2) positions are indicated. The numbers at the left indicate molecular mass in kilodaltons. (B) Calu3 cells were incubated with trypsin pretreated pps. Virus entry was quantified by measuring luciferase levels at 48-h posttransduction. Error bars present SD from the mean (n = 4). Statistical significance was assessed by Student's t test. ‡P < 0.001.
Fig. 4.
S1/S2 cleavage facilitates the receptor-induced exposure of a second S2′ cleavage site. (A) MERS (WT) pps were mixed with SFM or with DPP4 pps. The mixed pps were treated with increasing concentrations of trypsin. S and DPP4 proteins on pps were analyzed by Western blotting. Uncleaved (Sunc) and cleaved (S2, S2′, and 40K) positions are indicated. The numbers at the left indicate molecular mass in kilodaltons. (B) Similar experiments to A were carried out in parallel with YSAS MERS pps.
Fig. 5.
S1/S2 cleavage facilitates early virus entry. (A) Calu3 and Huh7 cells were incubated with protease inhibitors [100 µM camostat (camo), 50 µM PCI, 10 µM E64d], or with DMSO, and then transduced with MERS or VSV pps. (B) Three HAE cultures were incubated with indicated protease inhibitors, and then transduced with WT and S1/S2 mutant MERS pps. (C) Huh7 and Huh7/TMPRSS2 cells were incubated with protease inhibitors, and then transduced with WT or S1/S2 mutant (YSAS or SSVR) MERS pps. Virus entry was quantified by measuring luciferase levels in infected cells at 48-h (A, C) or 18-h (B) posttransduction. Error bars present SD from the mean (n = 3). Statistical significance was assessed by Student's t test. *P < 0.05; †P < 0.01; ‡P < 0.001; ns, not significant.
Fig. S1.
Effect of protease inhibitors on MERS pp entry into Caco2 and Vero81 cells. Caco2 and Vero81 cells were incubated with protease inhibitors [100 µM camostat (camo), 50 µM PCI, 10 µM E64d], or with DMSO. Cells then were transduced with MERS or VSV pps. Virus entry was quantified by measuring luciferase levels at 48 h posttransduction and plotted relative to DMSO control values. Error bars present SD from the mean (n = 3). Statistical significance was assessed by Student’s t test. *P < 0.05; ‡P < 0.001.
Fig. S2.
Effect of protease inhibitors on WT and mutant MERS pp entry into Caco2 cells. (A) Caco2 cells were incubated with WT, S1/S2 mutant (YSAS and SSVR) MERS pps or with pp-lacking S proteins (Bald). (B) Caco2 cells were incubated with protease inhibitors (100 µM camostat, 50 µM PCI, 10 µM E64d), or with DMSO. Cells were then transduced with WT or S1/S2 mutant (YSAS or SSVR) MERS pps. Virus entry was quantified by measuring luciferase levels at 48-h posttransduction and plotted relative to DMSO control values. Error bars present SD from the mean (n = 3). Statistical significance was assessed by Student's t test. †P < 0.01; ‡P < 0.001.
Fig. 6.
Uncleaved MERS entry depends on endosomal cathepsins. (A) DPP4, TMPRSS2, furin, cathepsin L (Cat. L), cathepsin B (Cat. B), and hypoxanthine phosphoribosyltransferase (HPRT) mRNA levels were measured in Huh7 cell extracts by quantitative RT-PCR, and plotted relative to HPRT. (B) The same mRNA levels were measured in Calu3 and HAE cell extracts and plotted relative to those in Huh7 cells. (C) Calu3 cells were transduced with lentiviral vectors encoding human cathepsin L or GFP. After 3 d, cells were treated with or without 10 µM E64d and then transduced with WT or S1/S2 mutant MERS pps. (D) Calu3 cells were incubated with WT or YSAS MERS pps at 4 °C, and then treated with cathepsin L. Virus entry was quantified by measuring luciferase levels at 48-h posttransduction. Error bars present SD from the mean (n = 3). Statistical significance was assessed by Student's t test. †P < 0.01.
Fig. 7.
MERS-CoV–cell entry model. In some producer cell types, MERS-CoV S proteins are cleaved by furin/proprotein convertases in the exocytic pathway. Cleaved MERS-CoV S proteins change their conformations rapidly after receptor binding, exposing subsequent proteolytic cleavage sites, which are processed by proteases (i.e., TMPRs, found at or near cell surfaces). Early cell-surface entry is achieved when several adjacent S proteins are processed. In other producer cell types, MERS-CoV S proteins are not cleaved. Uncleaved MERS-CoV S proteins slowly change their conformations after receptor binding. MERS-CoVs having uncleaved S proteins traffic to the late endosomes/lysosomes and late endosomal entry is achieved when several adjacent S proteins are eventually processed by cathepsins.
Similar articles
- The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases.
Earnest JT, Hantak MP, Li K, McCray PB Jr, Perlman S, Gallagher T. Earnest JT, et al. PLoS Pathog. 2017 Jul 31;13(7):e1006546. doi: 10.1371/journal.ppat.1006546. eCollection 2017 Jul. PLoS Pathog. 2017. PMID: 28759649 Free PMC article. - Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization.
Kleine-Weber H, Elzayat MT, Wang L, Graham BS, Müller MA, Drosten C, Pöhlmann S, Hoffmann M. Kleine-Weber H, et al. J Virol. 2019 Jan 4;93(2):e01381-18. doi: 10.1128/JVI.01381-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404801 Free PMC article. - Ca2+ Ions Promote Fusion of Middle East Respiratory Syndrome Coronavirus with Host Cells and Increase Infectivity.
Straus MR, Tang T, Lai AL, Flegel A, Bidon M, Freed JH, Daniel S, Whittaker GR. Straus MR, et al. J Virol. 2020 Jun 16;94(13):e00426-20. doi: 10.1128/JVI.00426-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295925 Free PMC article. - Coronavirus Spike Protein and Tropism Changes.
Hulswit RJ, de Haan CA, Bosch BJ. Hulswit RJ, et al. Adv Virus Res. 2016;96:29-57. doi: 10.1016/bs.aivir.2016.08.004. Epub 2016 Sep 13. Adv Virus Res. 2016. PMID: 27712627 Free PMC article. Review. - Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis.
Millet JK, Whittaker GR. Millet JK, et al. Virus Res. 2015 Apr 16;202:120-34. doi: 10.1016/j.virusres.2014.11.021. Epub 2014 Nov 22. Virus Res. 2015. PMID: 25445340 Free PMC article. Review.
Cited by
- Spike protein mutational landscape in India during the complete lockdown phase: Could Muller's ratchet be a future game-changer for COVID-19?
Banerjee R, Basak K, Ghosh A, Rajachandran V, Sureka K, Ganguly D, Chattopadhyay S. Banerjee R, et al. Infect Genet Evol. 2021 Aug;92:104874. doi: 10.1016/j.meegid.2021.104874. Epub 2021 Apr 24. Infect Genet Evol. 2021. PMID: 33905891 Free PMC article. - Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses.
Hu Y, Ma C, Szeto T, Hurst B, Tarbet B, Wang J. Hu Y, et al. ACS Infect Dis. 2021 Mar 12;7(3):586-597. doi: 10.1021/acsinfecdis.0c00761. Epub 2021 Mar 1. ACS Infect Dis. 2021. PMID: 33645977 Free PMC article. - Furin: A Potential Therapeutic Target for COVID-19.
Wu C, Zheng M, Yang Y, Gu X, Yang K, Li M, Liu Y, Zhang Q, Zhang P, Wang Y, Wang Q, Xu Y, Zhou Y, Zhang Y, Chen L, Li H. Wu C, et al. iScience. 2020 Oct 23;23(10):101642. doi: 10.1016/j.isci.2020.101642. Epub 2020 Oct 5. iScience. 2020. PMID: 33043282 Free PMC article. - Neutralizing antibodies against SARS-CoV-2: current understanding, challenge and perspective.
Huang Y, Sun H, Yu H, Li S, Zheng Q, Xia N. Huang Y, et al. Antib Ther. 2020 Dec 28;3(4):285-299. doi: 10.1093/abt/tbaa028. eCollection 2020 Dec. Antib Ther. 2020. PMID: 33912797 Free PMC article. Review. - Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy.
Zhang Q, Xiang R, Huo S, Zhou Y, Jiang S, Wang Q, Yu F. Zhang Q, et al. Signal Transduct Target Ther. 2021 Jun 11;6(1):233. doi: 10.1038/s41392-021-00653-w. Signal Transduct Target Ther. 2021. PMID: 34117216 Free PMC article. Review.
References
- Kido H, et al. Role of host cellular proteases in the pathogenesis of influenza and influenza-induced multiple organ failure. Biochim Biophys Acta. 2012;1824(1):186–194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI060699/AI/NIAID NIH HHS/United States
- P01 HL051670/HL/NHLBI NIH HHS/United States
- P01 HL091842/HL/NHLBI NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous