Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease - PubMed (original) (raw)
. 2018 Jan;67(1):108-119.
doi: 10.1136/gutjnl-2016-312135. Epub 2016 Oct 8.
Arnau Vich Vila 1 2, Marc Jan Bonder 2, Jingyuan Fu 3, Dirk Gevers 4, Marijn C Visschedijk 1 2, Lieke M Spekhorst 1 2, Rudi Alberts 1 2, Lude Franke 2, Hendrik M van Dullemen 1, Rinze W F Ter Steege 1, Curtis Huttenhower 4 5, Gerard Dijkstra 1, Ramnik J Xavier 4 6, Eleonora A M Festen 1 2, Cisca Wijmenga 2, Alexandra Zhernakova 2, Rinse K Weersma 1
Affiliations
- PMID: 27802154
- PMCID: PMC5699972
- DOI: 10.1136/gutjnl-2016-312135
Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease
Floris Imhann et al. Gut. 2018 Jan.
Abstract
Objective: Patients with IBD display substantial heterogeneity in clinical characteristics. We hypothesise that individual differences in the complex interaction of the host genome and the gut microbiota can explain the onset and the heterogeneous presentation of IBD. Therefore, we performed a case-control analysis of the gut microbiota, the host genome and the clinical phenotypes of IBD.
Design: Stool samples, peripheral blood and extensive phenotype data were collected from 313 patients with IBD and 582 truly healthy controls, selected from a population cohort. The gut microbiota composition was assessed by tag-sequencing the 16S rRNA gene. All participants were genotyped. We composed genetic risk scores from 11 functional genetic variants proven to be associated with IBD in genes that are directly involved in the bacterial handling in the gut: NOD2, CARD9, ATG16L1, IRGM and FUT2.
Results: Strikingly, we observed significant alterations of the gut microbiota of healthy individuals with a high genetic risk for IBD: the IBD genetic risk score was significantly associated with a decrease in the genus Roseburia in healthy controls (false discovery rate 0.017). Moreover, disease location was a major determinant of the gut microbiota: the gut microbiota of patients with colonic Crohn's disease (CD) is different from that of patients with ileal CD, with a decrease in alpha diversity associated to ileal disease (p=3.28×10-13).
Conclusions: We show for the first time that genetic risk variants associated with IBD influence the gut microbiota in healthy individuals. Roseburia spp are acetate-to-butyrate converters, and a decrease has already been observed in patients with IBD.
Keywords: BACTERIAL INTERACTIONS; GENETICS; INFLAMMATORY BOWEL DISEASE; INTESTINAL BACTERIA.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/.
Conflict of interest statement
Competing interests: None declared.
Figures
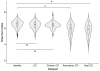
Figure 1
Alpha diversity (Shannon Index) of the gut microbiota of healthy controls, Ulcerative Colitis (UC) patients, colonic Crohn’s Disease (CD) patients, ileocolonic CD patients and ileal CD patients. Alpha diversity is not decreased in colonic disease (UC and colonic CD) compared to healthy controls. In contrast, in ileal and ileocolonic CD patients, the alpha diversity is statistically significantly decreased (ileal CD patients vs. healthy controls P = 3.28 × 10−13 and ileocolonic CD patients vs. healthy controls P = 3.11 × 10−11, Wilcoxon test).
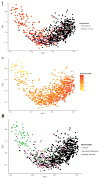
Figure 2
Principal Coordinate Analysis (PCoA) of stool samples of 313 IBD patients and 582 healthy controls. (A) The gut microbiota of IBD patients is different from the gut microbiota of healthy controls, with only partial overlap. (B) The first component is related to the Shannon Index. (C, D) There is more overlap between colonic disease (Ulcerative Colitis and colonic Crohn’s Disease combined) and healthy controls than between ileal disease (ileal Crohn’s Disease and ileocolonic Crohn’s Disease combined) and healthy controls. The first component is related to disease location (PCoA1 rho=0.63, P = 7.39 × 10−91, Spearman correlation) and colonic CD patients differ from ileal CD patients (P = 5.42 × 10−9).
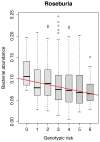
Figure 3
Increased risk score of 11 IBD related genetic variants in gut bacterial handling genes (NOD2, CARD9, IRGM, ATG16L1 and FUT2) is statistically significantly associated to decreased abundance of Roseburia spp. in healthy controls (FDR = 0.017).
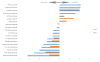
Figure 4
Fold change of increased and decreased bacterial families in UC and CD patients versus healthy controls (FDR < 0.05).
Comment in
- No association between urbanisation, neighbourhood deprivation and IBD: a population-based study of 4 million individuals.
Butwicka A, Sariaslan A, Larsson H, Halfvarson J, Myrelid PE, Olén O, Frisen L, Lichtenstein P, Ludvigsson JF. Butwicka A, et al. Gut. 2019 May;68(5):947-948. doi: 10.1136/gutjnl-2018-316326. Epub 2018 Apr 24. Gut. 2019. PMID: 29691277 No abstract available.
Similar articles
- Inflammatory bowel disease: between genetics and microbiota.
Younis N, Zarif R, Mahfouz R. Younis N, et al. Mol Biol Rep. 2020 Apr;47(4):3053-3063. doi: 10.1007/s11033-020-05318-5. Epub 2020 Feb 21. Mol Biol Rep. 2020. PMID: 32086718 Review. - The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection.
Stecher B. Stecher B. Microbiol Spectr. 2015 Jun;3(3). doi: 10.1128/microbiolspec.MBP-0008-2014. Microbiol Spectr. 2015. PMID: 26185088 - Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid.
Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Kabeerdoss J, et al. Indian J Med Res. 2015 Jul;142(1):23-32. doi: 10.4103/0971-5916.162091. Indian J Med Res. 2015. PMID: 26261163 Free PMC article. - Microbiota as Therapeutic Targets.
Xavier RJ. Xavier RJ. Dig Dis. 2016;34(5):558-65. doi: 10.1159/000445263. Epub 2016 Jun 22. Dig Dis. 2016. PMID: 27331403 - Determinants of IBD Heritability: Genes, Bugs, and More.
Turpin W, Goethel A, Bedrani L, Croitoru Mdcm K. Turpin W, et al. Inflamm Bowel Dis. 2018 May 18;24(6):1133-1148. doi: 10.1093/ibd/izy085. Inflamm Bowel Dis. 2018. PMID: 29701818 Free PMC article. Review.
Cited by
- Mesenchymal stem cells: a novel therapeutic approach for feline inflammatory bowel disease.
Xie Q, Gong S, Cao J, Li A, Kulyar MF, Wang B, Li J. Xie Q, et al. Stem Cell Res Ther. 2024 Nov 9;15(1):409. doi: 10.1186/s13287-024-04038-y. Stem Cell Res Ther. 2024. PMID: 39522034 Free PMC article. - The disturbance of intestinal microbiome caused by the novel duck reovirus infection in Cherry Valley ducklings can induce intestinal damage.
Li L, Lei B, Zhang W, Wang W, Shang C, Hu Y, Zhao K, Yuan W. Li L, et al. Poult Sci. 2024 Oct 19;103(12):104428. doi: 10.1016/j.psj.2024.104428. Online ahead of print. Poult Sci. 2024. PMID: 39490133 Free PMC article. - Gut microbial and human genetic signatures of inflammatory bowel disease increase risk of comorbid mental disorders.
Lee J, Oh SJ, Ha E, Shin GY, Kim HJ, Kim K, Lee CK. Lee J, et al. NPJ Genom Med. 2024 Oct 29;9(1):52. doi: 10.1038/s41525-024-00440-w. NPJ Genom Med. 2024. PMID: 39472439 Free PMC article. - The Gut Microbiome Advances Precision Medicine and Diagnostics for Inflammatory Bowel Diseases.
Mousa WK, Al Ali A. Mousa WK, et al. Int J Mol Sci. 2024 Oct 19;25(20):11259. doi: 10.3390/ijms252011259. Int J Mol Sci. 2024. PMID: 39457040 Free PMC article. Review. - Cardiovascular Disease May Be Triggered by Gut Microbiota, Microbial Metabolites, Gut Wall Reactions, and Inflammation.
Dicks LMT. Dicks LMT. Int J Mol Sci. 2024 Oct 2;25(19):10634. doi: 10.3390/ijms251910634. Int J Mol Sci. 2024. PMID: 39408963 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous