Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrin - PubMed (original) (raw)
Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrin
Julia Muenzner et al. Traffic. 2017 Jan.
Abstract
Short peptide motifs in unstructured regions of clathrin-adaptor proteins recruit clathrin to membranes to facilitate post-Golgi membrane transport. Three consensus clathrin-binding peptide sequences have been identified and structural studies show that each binds distinct sites on the clathrin heavy chain N-terminal domain (NTD). A fourth binding site for adaptors on NTD has been functionally identified but not structurally characterised. We have solved high resolution structures of NTD bound to peptide motifs from the cellular clathrin adaptors β2 adaptin and amphiphysin plus a putative viral clathrin adaptor, hepatitis D virus large antigen (HDAg-L). Surprisingly, with each peptide we observe simultaneous peptide binding at multiple sites on NTD and viral peptides binding to the same sites as cellular peptides. Peptides containing clathrin-box motifs (CBMs) with the consensus sequence LΦxΦ[DE] bind at the 'arrestin box' on NTD, between β-propeller blades 4 and 5, which had previously been thought to bind a distinct consensus sequence. Further, we structurally define the fourth peptide binding site on NTD, which we term the Royle box. In vitro binding assays show that clathrin is more readily captured by cellular CBMs than by HDAg-L, and site-directed mutagenesis confirms that multiple binding sites on NTD contribute to efficient capture by CBM peptides.
Short peptide motifs in unstructured regions of clathrin‐adaptor proteins recruit clathrin to membranes to facilitate post‐Golgi membrane transport. A total of 3 consensus clathrin‐binding peptide sequences have been identified and structural studies show that each binds distinct sites on the clathrin heavy chain N‐terminal domain (NTD). A fourth binding site for adaptors on NTD has been functionally identified but not structurally characterized. We have solved high‐resolution structures of NTD bound to peptide motifs from the cellular clathrin adaptors β2 adaptin and amphiphysin plus a putative viral clathrin adaptor, hepatitis D virus large antigen (HDAg‐L). Surprisingly, with each peptide we observe simultaneous peptide binding at multiple sites on NTD and viral peptides binding to the same sites as cellular peptides. Peptides containing clathrin‐box motifs (CBMs) with consensus sequence LΦxΦ[DE] bind at the “arrestin box” on NTD, between β‐propeller blades 4 and 5, which had previously been thought to bind a distinct consensus sequence. Further, we structurally define the fourth peptide binding site on NTD, which we term the Royle box. in vitro binding assays show that clathrin is more readily captured by cellular CBMs than by HDAg‐L, and site‐directed mutagenesis confirms that multiple binding sites on NTD contribute to efficient capture by CBM peptides.
Keywords: amphiphysin; arrestin; assembly polypeptide 2 (AP2); clathrin-mediated endocytosis; endocytosis; hepatitis D virus.
© 2016 The Authors. Traffic Published by John Wiley & Sons Ltd.
Figures
Figure 1
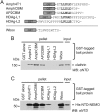
Cellular and viral peptide motifs bind clathrin N‐terminal domain (NTD) to different degrees. A, Glutathione S‐transferase (GST) fusions of clathrin‐binding peptides used in this study. Clathrin‐box motifs (CBMs) are aligned in bold. Amph4T1, human amphiphysin I CBM plus additional residues derived from the expression vector22; AmphCBM, human amphiphysin I CBM; AP2CBM, CBM from flexible hinge of β2 adaptin subunit of human AP2; HDAg‐L1, putative CBM from clade I hepatitis D virus large antigen; HDAg‐L2, putative CBM from clade II hepatitis D virus large antigen; Wbox, human amphiphysin W box binding motif. B, Capture (“GST pull‐down”) of purified clathrin by GST‐tagged clathrin‐binding peptides. Clathrin (input) was incubated with glutathione sepharose pre‐loaded with GST‐tagged “bait” proteins. After washing, proteins bound to the beads (pellet) were subjected to SDS‐PAGE and immunoblotting (WB) using an antibody that recognizes clathrin NTD (αNTD). C, Capture of His‐NTD‐NEMO by GST‐tagged clathrin binding peptides. Purified recombinant His‐NTD‐NEMO was used in GST pull‐down experiments as in (B).
Figure 2
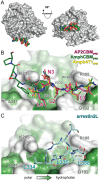
The clathrin‐box motifs (CBMs) of cellular and viral proteins bind multiple sites on clathrin N‐terminal domain (NTD). Unboxed image shows the β‐propeller fold of clathrin NTD (grey ribbons) with numbers enumerating the 7 β‐stranded blades. Spheres represent peptides bound at the 4 peptide‐interaction sites on NTD. Boxed images show CBM‐containing peptides (sticks, carbon atoms coloured as follows: AP2CBMpep, magenta; AmphCBMpep, dark green; Amph4T1pep, yellow; HDAg‐L1pep, orange; HDAg‐L2pep, light blue) bound at the clathrin box, arrestin box and Royle box sites on clathrin NTD (grey ribbons). Unbiased F O‐F C electron density (3 σ) used to place peptides into the structures is shown as a green mesh. Selected side chain atoms of clathrin NTD are shown (sticks with grey carbon atoms).
Figure 3
Cellular clathrin‐box motifs (CBMs) bind in a different conformation than arrestin2L at the arrestin box. A, The surface of clathrin N‐terminal domain (NTD) is shown (grey) oriented as in Figure 2 (left) and rotated by 90° around the horizontal axis (right). The AmphCBMpep peptide bound at the arrestin box is shown as coloured spheres. B, Close‐up view of cellular CBM‐containing peptides bound at the arrestin box. Peptides are shown as sticks coloured as in Figure 2. The surface of clathrin NTD is shown, coloured from high (green) to low (white) surface residue hydrophobicity, with outlines of selected surface side chains shown in grey. Bound AP2CBMpep residues are numbered by their position in the LΦxΦ[DE] CBM consensus sequence. C, The extended surface loop of arrestin 2 long isoform (arrestin2L) bound at the arrestin box (PDB 3GD1).13 NTD is shown as in (B) and arrestin2L residues 332‐340 are shown as sticks with cyan carbon atoms. Note that in (B) the direction of the bound peptides is right (N terminus) to left (C terminus), whereas in (C) the peptide chain between residues L334‐L338 runs in the opposite direction.
Figure 4
Localization and characterization of the fourth peptide binding site on clathrin N‐terminal domain (NTD): The Royle box. A, Amph4T1pep (left), HDAg‐L1pep (middle) and HDAg‐L2pep (right) peptides bound at the Royle box in feature‐enhanced maps24 calculated using the final refined model (2σ, magenta). For clarity, maps are shown only within 2 Å of the bound peptides. Peptides are shown as sticks, coloured as in Figure 2, and clathrin NTD is shown as a grey surface. B, The surface of clathrin NTD, coloured by amino acid conservation from conserved (magenta) to variable (blue), is shown oriented as in Figure 2 (left) and rotated by 90° around the vertical axis (right). The Amph4T1pep peptide bound at a conserved surface patch between NTD β‐propeller blades 6 and 7 (which we term the Royle box) is shown as sticks with yellow carbon atoms. C, Close‐up view of cellular and virus peptides bound at the Royle box. Peptides are shown as sticks coloured as in Figure 2. The surface of clathrin NTD is shown, coloured from high (green) to low (white) surface residue hydrophobicity, with outlines of selected surface side chains shown in grey. A hydrophobic NTD surface pocket that is occupied by hydrophobic residues of all three peptides is marked by a dotted line. The peptide sequences used for co‐crystallization are structurally aligned at the bottom of the panel. The directionality of the bound peptides is indicated. Residues that could be confidently modelled in the structures are highlighted and residues that form side chain interactions with NTD surface pockets are printed in bold type. D, Cellular and viral peptides bind near NTD residues functionally implicated in clathrin‐mediated endocytosis. The surface of NTD (grey) is oriented as in the right image of (A) with residues mutated by Willox and Royle14 (light and dark purple) or in this study (pink) highlighted. The Amph4T1pep peptide is shown as spheres. Inset shows the Amph4T1pep peptide (sticks with yellow carbon atoms) bound to NTD (grey, ribbon with selected side chains shown as sticks). The carbon atoms of residues substituted in clathrin mutants that disrupt transferrin uptake,14 a proxy for clathrin‐mediated endocytosis, are dark purple while those of residues where substitution does not affect transferrin uptake are light purple. The side chain of F9, mutated in this study, is coloured bright pink.
Figure 5
Mutation at single or multiple sites on clathrin N‐terminal domain (NTD) disrupts binding to peptide motifs. A, Ribbon representation of NTD (grey) showing the location of residues that were mutated on their own or in combination to disrupt peptide binding at the clathrin box (blue), arrestin box (green), W box (orange) and Royle box (purple). B, Thermal stability of single‐ or multiple‐site mutations of NTD as determined by differential scanning fluorimetry. The melting temperatures (TM) of mutants relative to that of wild‐type His‐NTD are shown (error bars represent the standard deviation of measurement in triplicate). Mutations that perturb the TM by more than 3 K (dotted line) were not considered further. C and D, Capture of NTD mutants by glutathione S‐transferase (GST)‐tagged clathrin‐binding peptides. Purified recombinant wild‐type or mutant His‐NTD‐NEMO was incubated with glutathione sepharose pre‐loaded with GST‐tagged “bait” proteins. After washing, proteins bound to the beads (pellet) were subjected to SDS‐PAGE and immunoblotting (WB) using an antibody that recognizes clathrin NTD (αNTD).
Figure 6
The overlapping β2 adaptin arrestin‐box and clathrin‐box motifs both bind multiple sites on clathrin N‐terminal domain (NTD). A, Glutathione S‐transferase (GST) fusions of the clathrin‐box motif (GST‐AP2CBM) and arrestin‐binding motif (GST‐AP2arrS and GST‐AP2arrL) from the hinge region of β2 adaptin, the arrestin‐box motif constructs having either the next residue of β2 adaptin (“L”, GST‐AP2arrS) or the sequence that follows the LLGDL motif of arrestin2L (“ASS”, GST‐AP2arrL) appended at their C termini. B, Capture of wild‐type NTD or a mutant with disrupted clathrin and Royle boxes (Q89A + F91K + F9W) by GST‐tagged β2 adaptin clathrin‐binding motifs. Purified recombinant wild‐type or mutant His‐NTD‐NEMO was incubated with glutathione sepharose pre‐loaded with GST‐tagged “bait” proteins. After washing, proteins bound to the beads (pellet) were subjected to SDS‐PAGE and immunoblotting (WB) using an antibody that recognizes clathrin NTD (αNTD).
Similar articles
- Identification of a novel interaction site between the large hepatitis delta antigen and clathrin that regulates the assembly of genotype III hepatitis delta virus.
Chiou WC, Lu HF, Chen JC, Lai YH, Chang MF, Huang YL, Tien N, Huang C. Chiou WC, et al. Virol J. 2022 Oct 17;19(1):163. doi: 10.1186/s12985-022-01866-3. Virol J. 2022. PMID: 36253859 Free PMC article. - Clathrin functions in the absence of the terminal domain binding site for adaptor-associated clathrin-box motifs.
Collette JR, Chi RJ, Boettner DR, Fernandez-Golbano IM, Plemel R, Merz AJ, Geli MI, Traub LM, Lemmon SK. Collette JR, et al. Mol Biol Cell. 2009 Jul;20(14):3401-13. doi: 10.1091/mbc.e08-10-1082. Epub 2009 May 20. Mol Biol Cell. 2009. PMID: 19458198 Free PMC article. - Clathrin-mediated post-Golgi membrane trafficking in the morphogenesis of hepatitis delta virus.
Huang C, Chang SC, Yang HC, Chien CL, Chang MF. Huang C, et al. J Virol. 2009 Dec;83(23):12314-24. doi: 10.1128/JVI.01044-09. Epub 2009 Sep 30. J Virol. 2009. PMID: 19793827 Free PMC article. - ENTH/ANTH proteins and clathrin-mediated membrane budding.
Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. Legendre-Guillemin V, et al. J Cell Sci. 2004 Jan 1;117(Pt 1):9-18. doi: 10.1242/jcs.00928. J Cell Sci. 2004. PMID: 14657269 Review. - Getting in touch with the clathrin terminal domain.
Lemmon SK, Traub LM. Lemmon SK, et al. Traffic. 2012 Apr;13(4):511-9. doi: 10.1111/j.1600-0854.2011.01321.x. Epub 2012 Jan 13. Traffic. 2012. PMID: 22239657 Free PMC article. Review.
Cited by
- Clathrin's adaptor interaction sites are repurposed to stabilize microtubules during mitosis.
Rondelet A, Lin YC, Singh D, Porfetye AT, Thakur HC, Hecker A, Brinkert P, Schmidt N, Bendre S, Müller F, Mazul L, Widlund PO, Bange T, Hiller M, Vetter IR, Bird AW. Rondelet A, et al. J Cell Biol. 2020 Feb 3;219(2):e201907083. doi: 10.1083/jcb.201907083. J Cell Biol. 2020. PMID: 31932847 Free PMC article. - Extreme Fuzziness: Direct Interactions between Two IDPs.
Wang W, Wang D. Wang W, et al. Biomolecules. 2019 Feb 26;9(3):81. doi: 10.3390/biom9030081. Biomolecules. 2019. PMID: 30813629 Free PMC article. Review. - Identification of a novel interaction site between the large hepatitis delta antigen and clathrin that regulates the assembly of genotype III hepatitis delta virus.
Chiou WC, Lu HF, Chen JC, Lai YH, Chang MF, Huang YL, Tien N, Huang C. Chiou WC, et al. Virol J. 2022 Oct 17;19(1):163. doi: 10.1186/s12985-022-01866-3. Virol J. 2022. PMID: 36253859 Free PMC article. - Role of Clathrin and Dynamin in Clathrin Mediated Endocytosis/Synaptic Vesicle Recycling and Implications in Neurological Diseases.
Prichard KL, O'Brien NS, Murcia SR, Baker JR, McCluskey A. Prichard KL, et al. Front Cell Neurosci. 2022 Jan 18;15:754110. doi: 10.3389/fncel.2021.754110. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35115907 Free PMC article. Review. - Behaviour of intrinsically disordered proteins in protein-protein complexes with an emphasis on fuzziness.
Olsen JG, Teilum K, Kragelund BB. Olsen JG, et al. Cell Mol Life Sci. 2017 Sep;74(17):3175-3183. doi: 10.1007/s00018-017-2560-7. Epub 2017 Jun 8. Cell Mol Life Sci. 2017. PMID: 28597296 Free PMC article. Review.
References
- Unanue ER, Ungewickell E, Branton D. The binding of clathrin triskelions to membranes from coated vesicles. Cell. 1981;26(3 Pt 1):439‐446. - PubMed
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699‐727. - PubMed
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. 2004;20:153‐191. - PubMed
- Shih W, Gallusser A, Kirchhausen T. A clathrin‐binding site in the hinge of the beta 2 chain of mammalian AP‐2 complexes. J Biol Chem. 1995;270(52):31083‐31090. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials