Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors - PubMed (original) (raw)
Review
Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors
Muhammad A Zabidi et al. Trends Genet. 2016 Dec.
Abstract
Gene expression is regulated by genomic enhancers that recruit transcription factors and cofactors to activate transcription from target core promoters. Over the past years, thousands of enhancers and core promoters in animal genomes have been annotated, and we have learned much about the domain structure in which regulatory genomes are organized in animals. Enhancer-core-promoter targeting occurs at several levels, including regulatory domains, DNA accessibility, and sequence-encoded core-promoter specificities that are likely mediated by different regulatory proteins. We review here current knowledge about enhancer-core-promoter targeting, regulatory communication between enhancers and core promoters, and the protein factors involved. We conclude with an outlook on open questions that we find particularly interesting and that will likely lead to additional insights in the upcoming years.
Keywords: cofactors; core promoters; enhancers; enhancer–core-promoter specificity; transcription factors; transcription regulation.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
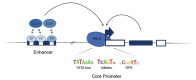
Figure 1. Enhancers and core-promoters, two major classes of _cis-_regulatory elements.
An enhancer contains binding sites for sequence-specific transcription factors (TFs). These in turn recruit cofactors (COFs) that typically mediate the regulatory communication between the core promoter and the enhancer, i.e. relay the enhancer’s regulatory cues to the enhancer’s target core promoters. Core-promoters encompass short sequences of ~100bp surrounding the transcription start site, where Polymerase II (Pol II) assembles and initiates transcription. Core-promoters typically contain characteristic core-promoter elements or motifs, for example TATA box, Initiator or Downstream Promoter Element (DPE).
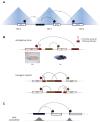
Figure 2. Topologically associating domains (TADs), promoter-proximal tethering elements (PTEs), and DNA accessibility.
(A) Enhancer function is typically restricted to activate core-promoters within the same TADs [12,31], the boundaries of which are enriched in insulator protein binding. (B) PTEs are sequences proximal to core-promoters that promote the preferential interaction between enhancers and core-promoters [–39]. For example, the PTE of the Sex combs reduced (Scr) core-promoter enables its activation by the distally-located Scr enhancer, skipping the intervening fushi tarazu (ftz). In transgenic reporter, relocation of the PTE to a proximal position of the ftz core-promoter results in activation of the latter by the Scr enhancer. In situ staining of embryo images are from Berkeley Drosophila Genome Project [161]. (C) Inaccessible DNA precludes a _cis-_regulatory element from participating in potential interaction. For example, Drosophila enhancers skip the proximal and inaccessible core-promoters, to activate more distal and accessible core-promoters [24].
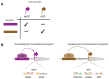
Figure 3. Sequence-mediated enhancer–core-promoter specificity.
(A) housekeeping core promoters (hkCP; purple) show preferences towards housekeeping over developmental enhancers. The reverse is true for developmental core promoters (dCP; ochre; ref [60]). (B) housekeeping enhancers bind DREF and activate transcription from hkCP, which typically contain Ohler Motifs 1 and 6 core-promoter elements (indicated by motif logos). Developmental enhancers meanwhile bind Trl and activate transcription from dCPs that contain different core-promoter elements, such as TATA box, Initiator and DPE. Compared to housekeeping enhancers which are TSS-proximal or even overlapping, developmental enhancers are found at various positions, including within introns or very distal.
Figure 4. Trans factors mediate sequence-mediated enhancer–core-promoter specificity.
(A) Activator bypass experiments show that different transcription factors (TFs) and cofactors (COFs) can differentially active hkCP over dCP. Activation is indicated by a check mark and examples of TFs and cofactors that function accordingly are shown below. (B) hkCP and dCP recruit TRF2- and TBP-containing complexes, respectively. The members of these complexes also potentially differ.
Figure 5. Enhancers activate core-promoters in a microenvironment.
(A) Static model of transcription regulation in which defined protein complexes formed by static protein-protein interactions at enhancers exert their function on core promoters. This model is incompatible with some observations such as the simultaneous activation of two core-promoters by a single enhancer [123]. (B) Transcription regulation might alternatively occur via an activating microenvironment, in which enhancers and core-promoters recruit trans factors to create a high concentration of regulatory proteins that dynamically interact with each other, and enable regulatory communication through post-transcription modifications (PTMs, top) or dynamic protein-protein-interactions and recruitment (bottom).
Similar articles
- Combinatorial function of transcription factors and cofactors.
Reiter F, Wienerroither S, Stark A. Reiter F, et al. Curr Opin Genet Dev. 2017 Apr;43:73-81. doi: 10.1016/j.gde.2016.12.007. Epub 2017 Jan 19. Curr Opin Genet Dev. 2017. PMID: 28110180 Review. - Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation.
Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Zabidi MA, et al. Nature. 2015 Feb 26;518(7540):556-9. doi: 10.1038/nature13994. Epub 2014 Dec 15. Nature. 2015. PMID: 25517091 Free PMC article. - Dynamic modulation of enhancer-promoter and promoter-promoter connectivity in gene regulation.
Makino S, Fukaya T. Makino S, et al. Bioessays. 2024 Sep;46(9):e2400101. doi: 10.1002/bies.202400101. Epub 2024 Jun 25. Bioessays. 2024. PMID: 38922969 Review. - Insights into gene regulation: From regulatory genomic elements to DNA-protein and protein-protein interactions.
Serebreni L, Stark A. Serebreni L, et al. Curr Opin Cell Biol. 2021 Jun;70:58-66. doi: 10.1016/j.ceb.2020.11.009. Epub 2020 Dec 29. Curr Opin Cell Biol. 2021. PMID: 33385708 Review. - Evaluating Enhancer Function and Transcription.
Field A, Adelman K. Field A, et al. Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review.
Cited by
- Identification and prediction of developmental enhancers in sea urchin embryos.
Arenas-Mena C, Miljovska S, Rice EJ, Gurges J, Shashikant T, Wang Z, Ercan S, Danko CG. Arenas-Mena C, et al. BMC Genomics. 2021 Oct 19;22(1):751. doi: 10.1186/s12864-021-07936-0. BMC Genomics. 2021. PMID: 34666684 Free PMC article. - Mutations in ACTRT1 and its enhancer RNA elements lead to aberrant activation of Hedgehog signaling in inherited and sporadic basal cell carcinomas.
Bal E, Park HS, Belaid-Choucair Z, Kayserili H, Naville M, Madrange M, Chiticariu E, Hadj-Rabia S, Cagnard N, Kuonen F, Bachmann D, Huber M, Le Gall C, Côté F, Hanein S, Rosti RÖ, Aslanger AD, Waisfisz Q, Bodemer C, Hermine O, Morice-Picard F, Labeille B, Caux F, Mazereeuw-Hautier J, Philip N, Levy N, Taieb A, Avril MF, Headon DJ, Gyapay G, Magnaldo T, Fraitag S, Crollius HR, Vabres P, Hohl D, Munnich A, Smahi A. Bal E, et al. Nat Med. 2017 Oct;23(10):1226-1233. doi: 10.1038/nm.4368. Epub 2017 Sep 4. Nat Med. 2017. PMID: 28869610 - KnockTF 2.0: a comprehensive gene expression profile database with knockdown/knockout of transcription (co-)factors in multiple species.
Feng C, Song C, Song S, Zhang G, Yin M, Zhang Y, Qian F, Wang Q, Guo M, Li C. Feng C, et al. Nucleic Acids Res. 2024 Jan 5;52(D1):D183-D193. doi: 10.1093/nar/gkad1016. Nucleic Acids Res. 2024. PMID: 37956336 Free PMC article. - Cis-Regulatory Elements in Mammals.
Liu X, Chen M, Qu X, Liu W, Dou Y, Liu Q, Shi D, Jiang M, Li H. Liu X, et al. Int J Mol Sci. 2023 Dec 26;25(1):343. doi: 10.3390/ijms25010343. Int J Mol Sci. 2023. PMID: 38203513 Free PMC article. Review. - The regulatory genome of the malaria vector Anopheles gambiae: integrating chromatin accessibility and gene expression.
Ruiz JL, Ranford-Cartwright LC, Gómez-Díaz E. Ruiz JL, et al. NAR Genom Bioinform. 2021 Jan 20;3(1):lqaa113. doi: 10.1093/nargab/lqaa113. eCollection 2021 Mar. NAR Genom Bioinform. 2021. PMID: 33987532 Free PMC article.
References
- Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. - PubMed
- Arrowsmith CH, et al. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. - PubMed
- Dawson MA, Kouzarides T. Cancer Epigenetics: From Mechanism to Therapy. Cell. 2012;150:12–27. - PubMed
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends in Biochemical Sciences. 1996;21:327–335. - PubMed
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous