Piphillin: Improved Prediction of Metagenomic Content by Direct Inference from Human Microbiomes - PubMed (original) (raw)
Piphillin: Improved Prediction of Metagenomic Content by Direct Inference from Human Microbiomes
Shoko Iwai et al. PLoS One. 2016.
Abstract
Functional analysis of a clinical microbiome facilitates the elucidation of mechanisms by which microbiome perturbation can cause a phenotypic change in the patient. The direct approach for the analysis of the functional capacity of the microbiome is via shotgun metagenomics. An inexpensive method to estimate the functional capacity of a microbial community is through collecting 16S rRNA gene profiles then indirectly inferring the abundance of functional genes. This inference approach has been implemented in the PICRUSt and Tax4Fun software tools. However, those tools have important limitations since they rely on outdated functional databases and uncertain phylogenetic trees and require very specific data pre-processing protocols. Here we introduce Piphillin, a straightforward algorithm independent of any proposed phylogenetic tree, leveraging contemporary functional databases and not obliged to any singular data pre-processing protocol. When all three inference tools were evaluated against actual shotgun metagenomics, Piphillin was superior in predicting gene composition in human clinical samples compared to both PICRUSt and Tax4Fun (p<0.01 and p<0.001, respectively) and Piphillin's ability to predict disease associations with specific gene orthologs exhibited a 15% increase in balanced accuracy compared to PICRUSt. From laboratory animal samples, no performance advantage was observed for any one of the tools over the others and for environmental samples all produced unsatisfactory predictions. Our results demonstrate that functional inference using the direct method implemented in Piphillin is preferable for clinical biospecimens. Piphillin is publicly available for academic use at http://secondgenome.com/Piphillin.
Conflict of interest statement
This work was supported in part by Second Genome Inc and Allergan PLC. Neil Poloso is employed by Allergen PLC and holds stock options. Shoko Iwai, Thomas Weinmaier, Karim Dabbagh, and Todd DeSantis are employed by Second Genome Inc. and hold stock options. Both Allergan PLC and Second Genome Inc. are independent therapeutics companies with products in development to treat gastrointestinal disorders and other human diseases. A publication announcing the availability of PiPhillin analysis for academic use will not affect the value of our therapeutic products. There are no PiPhillin patents, products in development or marketed products to declare. Second Genome, Inc. provides a commercial microbiome profiling service using software with demonstrable accuracy such as PiPhillin. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Figures
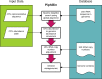
Fig 1. Piphillin algorithm.
The representative sequence of each OTU in the sample is first searched against 16S rRNA sequences in the genome database to obtain inferred genome(s). Then the OTU abundance table is converted to a genome abundance table. The resulting table is normalized by the 16S rRNA copy number of each genome and a metagenome is inferred using the gene contents (copy number of each gene) of each genome in the database.
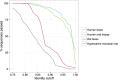
Fig 2. 16S rRNA gene amplicon sequences passing the identity threshold to the reference genomes.
Percentage of amplicon sequences from three datasets passing identity cutoffs from 0.75 to 1.00 against 16S rRNA gene sequences in the genome database were depicted. Green line, human feces dataset; blue line, human oral biopsy dataset; pink line, rat feces dataset; gray line, hypersaline microbial mat dataset.
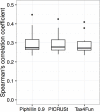
Fig 3. Spearman’s correlation coefficient between Piphillin results and shotgun metagenomics at ten different identity cutoffs tested in Piphillin.
Spearman’s correlation coefficient was calculated for each sample and mean, 1st and 3rd quartiles are depicted by the boxes. Whiskers extend to the furthest points within 150% of the interquartile range. Green, human feces dataset; blue, human oral biopsy dataset; pink rat feces dataset; gray, hypersaline microbial mat dataset.
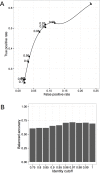
Fig 4. Sensitivity and specificity in identifying differentially abundant KOs from Piphillin against corresponding metagenomics.
(A) True positive rate and false positive rate of detecting significantly differentially abundant KOs in human oral biopsy sample. Numbers next to each point represent identity cutoff used for Piphillin. (B) Balanced accuracy of Piphillin at each identity cutoff.
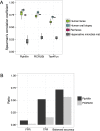
Fig 5. Comparison between Piphillin, PICRUSt and Tax4Fun.
(A) Spearman’s correlation coefficient against corresponding shotgun metagenomics results were compared. Spearman’s correlation coefficient was calculated for each sample and ranges are depicted as box and whisker plots as described in Fig 3. Green, human feces dataset; blue, human oral biopsy dataset; pink rat feces dataset; gray, hypersaline microbial mat dataset. (B) False positive rate, true positive rate and balanced accuracy of detecting significant differences between cancer and healthy human oral biopsy samples were compared.
Fig 6. Comparison between Piphillin with 0.9 identity cutoff and two other approaches.
Spearman’s correlation coefficient against shotgun metagenomics results was calculated for hypersaline microbial mat dataset. Ranges are depicted as box and whisker plots as described in Fig 3.
Similar articles
- Piphillin predicts metagenomic composition and dynamics from DADA2-corrected 16S rDNA sequences.
Narayan NR, Weinmaier T, Laserna-Mendieta EJ, Claesson MJ, Shanahan F, Dabbagh K, Iwai S, DeSantis TZ. Narayan NR, et al. BMC Genomics. 2020 Jan 17;21(1):56. doi: 10.1186/s12864-019-6427-1. BMC Genomics. 2020. PMID: 31952477 Free PMC article. - Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories.
Sun S, Jones RB, Fodor AA. Sun S, et al. Microbiome. 2020 Apr 2;8(1):46. doi: 10.1186/s40168-020-00815-y. Microbiome. 2020. PMID: 32241293 Free PMC article. - Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data.
Aßhauer KP, Wemheuer B, Daniel R, Meinicke P. Aßhauer KP, et al. Bioinformatics. 2015 Sep 1;31(17):2882-4. doi: 10.1093/bioinformatics/btv287. Epub 2015 May 7. Bioinformatics. 2015. PMID: 25957349 Free PMC article. - Use of whole genome shotgun metagenomics: a practical guide for the microbiome-minded physician scientist.
Ma J, Prince A, Aagaard KM. Ma J, et al. Semin Reprod Med. 2014 Jan;32(1):5-13. doi: 10.1055/s-0033-1361817. Epub 2014 Jan 3. Semin Reprod Med. 2014. PMID: 24390915 Review. - Metagenomic approaches: effective tools for monitoring the structure and functionality of microbiomes in anaerobic digestion systems.
Carabeo-Pérez A, Guerra-Rivera G, Ramos-Leal M, Jiménez-Hernández J. Carabeo-Pérez A, et al. Appl Microbiol Biotechnol. 2019 Dec;103(23-24):9379-9390. doi: 10.1007/s00253-019-10052-5. Epub 2019 Nov 9. Appl Microbiol Biotechnol. 2019. PMID: 31420693 Review.
Cited by
- Decoding microbial genomes to understand their functional roles in human complex diseases.
Wang Y, Dong Q, Hu S, Zou H, Wu T, Shi J, Zhang H, Sheng Y, Sun W, Kong X, Chen L. Wang Y, et al. Imeta. 2022 Mar 29;1(2):e14. doi: 10.1002/imt2.14. eCollection 2022 Jun. Imeta. 2022. PMID: 38868571 Free PMC article. Review. - Characterization of the gut bacterial and viral microbiota in latent autoimmune diabetes in adults.
Poulsen CS, Hesse D, Fernandes GR, Hansen TH, Kern T, Linneberg A, Van Espen L, Jørgensen T, Nielsen T, Alibegovic AC, Matthijnssens J, Pedersen O, Vestergaard H, Hansen T, Andersen MK. Poulsen CS, et al. Sci Rep. 2024 Apr 9;14(1):8315. doi: 10.1038/s41598-024-58985-w. Sci Rep. 2024. PMID: 38594375 Free PMC article. - Structural and Functional Shifts in the Microbial Community of a Heavy Metal-Contaminated Soil Exposed to Short-Term Changes in Air Temperature, Soil Moisture and UV Radiation.
Silva I, Alves M, Malheiro C, Silva ARR, Loureiro S, Henriques I, González-Alcaraz MN. Silva I, et al. Genes (Basel). 2024 Jan 16;15(1):107. doi: 10.3390/genes15010107. Genes (Basel). 2024. PMID: 38254996 Free PMC article. - Dry Stamping Coral Powder: An Effective Method for Isolating Coral Symbiotic Actinobacteria.
Becerril-Espinosa A, Mateos-Salmón C, Burgos A, Rodríguez-Zaragoza FA, Meza-Canales ID, Juarez-Carrillo E, Rios-Jara E, Ocampo-Alvarez H. Becerril-Espinosa A, et al. Microorganisms. 2023 Dec 10;11(12):2951. doi: 10.3390/microorganisms11122951. Microorganisms. 2023. PMID: 38138095 Free PMC article. - Effect of Red-Beetroot-Supplemented Diet on Gut Microbiota Composition and Metabolite Profile of Weaned Pigs-A Pilot Study.
Adekolurejo OO, McDermott K, Greathead HMR, Miller HM, Mackie AR, Boesch C. Adekolurejo OO, et al. Animals (Basel). 2023 Jul 4;13(13):2196. doi: 10.3390/ani13132196. Animals (Basel). 2023. PMID: 37443994 Free PMC article.
References
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105: 16731–6. 10.1073/pnas.0804812105 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources