Trans-ethnic follow-up of breast cancer GWAS hits using the preferential linkage disequilibrium approach - PubMed (original) (raw)
Multicenter Study
. 2016 Dec 13;7(50):83160-83176.
doi: 10.18632/oncotarget.13075.
Lori Shepherd 1, Kathryn L Lunetta 2, Song Yao 3, Qian Liu 1, Qiang Hu 1, Stephen A Haddad 4, Lara Sucheston-Campbell 3, Jeannette T Bensen 5, Elisa V Bandera 6, Lynn Rosenberg 4, Song Liu 1, Christopher A Haiman 7, Andrew F Olshan 5, Julie R Palmer 4, Christine B Ambrosone 3
Affiliations
- PMID: 27825120
- PMCID: PMC5341253
- DOI: 10.18632/oncotarget.13075
Multicenter Study
Trans-ethnic follow-up of breast cancer GWAS hits using the preferential linkage disequilibrium approach
Qianqian Zhu et al. Oncotarget. 2016.
Abstract
Leveraging population-distinct linkage equilibrium (LD) patterns, trans-ethnic follow-up of variants discovered from genome-wide association studies (GWAS) has proved to be useful in facilitating the identification of bona fide causal variants. We previously developed the preferential LD approach, a novel method that successfully identified causal variants driving the GWAS signals within European-descent populations even when the causal variants were only weakly linked with the GWAS-discovered variants. To evaluate the performance of our approach in a trans-ethnic setting, we applied it to follow up breast cancer GWAS hits identified mostly from populations of European ancestry in African Americans (AA). We evaluated 74 breast cancer GWAS variants in 8,315 AA women from the African American Breast Cancer Epidemiology and Risk (AMBER) consortium. Only 27% of them were associated with breast cancer risk at significance level α=0.05, suggesting race-specificity of the identified breast cancer risk loci. We followed up on those replicated GWAS hits in the AMBER consortium utilizing the preferential LD approach, to search for causal variants or better breast cancer markers from the 1000 Genomes variant catalog. Our approach identified stronger breast cancer markers for 80% of the GWAS hits with at least nominal breast cancer association, and in 81% of these cases, the marker identified was among the top 10 of all 1000 Genomes variants in the corresponding locus. The results support trans-ethnic application of the preferential LD approach in search for candidate causal variants, and may have implications for future genetic research of breast cancer in AA women.
Keywords: causal variant; fine-mapping; genome-wide association studies.
Conflict of interest statement
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Figures
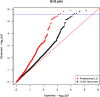
Figure 1. The QQ plot of overall breast cancer association p-values in AMBER consortium
The variants selected by the preferential LD approach in the 18 replicated loci are in red. The 1000 Genomes variants in the same 18 loci are in black. The blue horizontal line corresponds to the study-wide significance cutoff 2.55×10−6.
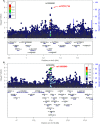
Figure 2. Breast cancer association of variants within 500 kb of rs10069690
A. and rs2363956 B. in the AMBER cohort. The GWAS-discovered variants were denoted by the purple circles.
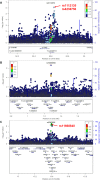
Figure 3. Association between variants within 500 kb of rs3112572 and ER+ breast cancer
A., between variants within 500 kb of rs10069690 B. and rs2363956 C. and ER- breast cancer in the AMBER cohort. The GWAS-discovered variants were denoted by the purple circles.
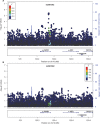
Figure 4. Breast cancer association of variants within 500 kb of rs2981582 in the AMBER cohort
The GWAS-discovered variant rs2981582 is denoted by the purple circle. The -logP values before A. and after B. conditioning on the causal variant rs2981578 were shown.
Similar articles
- Fine mapping of breast cancer genome-wide association studies loci in women of African ancestry identifies novel susceptibility markers.
Zheng Y, Ogundiran TO, Falusi AG, Nathanson KL, John EM, Hennis AJ, Ambs S, Domchek SM, Rebbeck TR, Simon MS, Nemesure B, Wu SY, Leske MC, Odetunde A, Niu Q, Zhang J, Afolabi C, Gamazon ER, Cox NJ, Olopade CO, Olopade OI, Huo D. Zheng Y, et al. Carcinogenesis. 2013 Jul;34(7):1520-8. doi: 10.1093/carcin/bgt090. Epub 2013 Mar 8. Carcinogenesis. 2013. PMID: 23475944 Free PMC article. - Genetic variants on chromosome 5p12 are associated with risk of breast cancer in African American women: the Black Women's Health Study.
Ruiz-Narvaez EA, Rosenberg L, Rotimi CN, Cupples LA, Boggs DA, Adeyemo A, Cozier YC, Adams-Campbell LL, Palmer JR. Ruiz-Narvaez EA, et al. Breast Cancer Res Treat. 2010 Sep;123(2):525-30. doi: 10.1007/s10549-010-0775-5. Epub 2010 Feb 7. Breast Cancer Res Treat. 2010. PMID: 20140701 Free PMC article. - Replication of breast cancer susceptibility loci in whites and African Americans using a Bayesian approach.
O'Brien KM, Cole SR, Poole C, Bensen JT, Herring AH, Engel LS, Millikan RC. O'Brien KM, et al. Am J Epidemiol. 2014 Feb 1;179(3):382-94. doi: 10.1093/aje/kwt258. Epub 2013 Nov 10. Am J Epidemiol. 2014. PMID: 24218030 Free PMC article. - From candidate gene studies to GWAS and post-GWAS analyses in breast cancer.
Fachal L, Dunning AM. Fachal L, et al. Curr Opin Genet Dev. 2015 Feb;30:32-41. doi: 10.1016/j.gde.2015.01.004. Epub 2015 Feb 27. Curr Opin Genet Dev. 2015. PMID: 25727315 Review. - Accounting for linkage disequilibrium in association analysis of diverse populations.
Charles BA, Shriner D, Rotimi CN. Charles BA, et al. Genet Epidemiol. 2014 Apr;38(3):265-73. doi: 10.1002/gepi.21788. Epub 2014 Jan 26. Genet Epidemiol. 2014. PMID: 24464495 Review.
Cited by
- Demographic, lifestyle, and genetic determinants of circulating concentrations of 25-hydroxyvitamin D and vitamin D-binding protein in African American and European American women.
Yao S, Hong CC, Bandera EV, Zhu Q, Liu S, Cheng TD, Zirpoli G, Haddad SA, Lunetta KL, Ruiz-Narvaez EA, McCann SE, Troester MA, Rosenberg L, Palmer JR, Olshan AF, Ambrosone CB. Yao S, et al. Am J Clin Nutr. 2017 Jun;105(6):1362-1371. doi: 10.3945/ajcn.116.143248. Epub 2017 Apr 19. Am J Clin Nutr. 2017. PMID: 28424184 Free PMC article. - Investigation of triple-negative breast cancer risk alleles in an International African-enriched cohort.
Martini R, Chen Y, Jenkins BD, Elhussin IA, Cheng E, Hoda SA, Ginter PS, Hanover J, Zeidan RB, Oppong JK, Adjei EK, Jibril A, Chitale D, Bensenhaver JM, Awuah B, Bekele M, Abebe E, Kyei I, Aitpillah FS, Adinku MO, Ankomah K, Osei-Bonsu EB, Nathansan SD, Jackson L, Jiagge E, Petersen LF, Proctor E, Nikolinakos P, Gyan KK, Yates C, Kittles R, Newman LA, Davis MB. Martini R, et al. Sci Rep. 2021 Apr 29;11(1):9247. doi: 10.1038/s41598-021-88613-w. Sci Rep. 2021. PMID: 33927264 Free PMC article. - Atypical Chemokine Receptor 1 (DARC/ACKR1) in Breast Tumors Is Associated with Survival, Circulating Chemokines, Tumor-Infiltrating Immune Cells, and African Ancestry.
Jenkins BD, Martini RN, Hire R, Brown A, Bennett B, Brown I, Howerth EW, Egan M, Hodgson J, Yates C, Kittles R, Chitale D, Ali H, Nathanson D, Nikolinakos P, Newman L, Monteil M, Davis MB. Jenkins BD, et al. Cancer Epidemiol Biomarkers Prev. 2019 Apr;28(4):690-700. doi: 10.1158/1055-9965.EPI-18-0955. Cancer Epidemiol Biomarkers Prev. 2019. PMID: 30944146 Free PMC article. - Early-onset triple-negative breast cancer in multiracial/ethnic populations: Distinct trends of prevalence of truncation mutations.
Liu Q, Yao S, Zhao H, Hu Q, Kwan ML, Roh JM, Ambrosone CB, Kushi LH, Liu S, Zhu Q. Liu Q, et al. Cancer Med. 2019 Apr;8(4):1845-1853. doi: 10.1002/cam4.2047. Epub 2019 Mar 12. Cancer Med. 2019. PMID: 30864286 Free PMC article. - An Interactive Resource to Probe Genetic Diversity and Estimated Ancestry in Cancer Cell Lines.
Dutil J, Chen Z, Monteiro AN, Teer JK, Eschrich SA. Dutil J, et al. Cancer Res. 2019 Apr 1;79(7):1263-1273. doi: 10.1158/0008-5472.CAN-18-2747. Epub 2019 Mar 20. Cancer Res. 2019. PMID: 30894373 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA179715/CA/NCI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- UM1 CA164973/CA/NCI NIH HHS/United States
- P30 CA016056/CA/NCI NIH HHS/United States
- R01 CA098663/CA/NCI NIH HHS/United States
- P01 CA151135/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- P50 CA058223/CA/NCI NIH HHS/United States
- R01 CA058420/CA/NCI NIH HHS/United States
- UM1 CA164974/CA/NCI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- P2C HD050924/HD/NICHD NIH HHS/United States
- R01 CA185623/CA/NCI NIH HHS/United States
- R01 CA100598/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials