Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform - PubMed (original) (raw)
. 2016 Nov 17;167(5):1215-1228.e25.
doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10.
Seung Joong Kim 2, Yi Shi 3, Paula Upla 4, Riccardo Pellarin 5, Michael Gagnon 6, Ilan E Chemmama 2, Junjie Wang 3, Ilona Nudelman 1, Wenzhu Zhang 3, Rosemary Williams 1, William J Rice 7, David L Stokes 4, Daniel Zenklusen 6, Brian T Chait 8, Andrej Sali 9, Michael P Rout 10
Affiliations
- PMID: 27839866
- PMCID: PMC5130164
- DOI: 10.1016/j.cell.2016.10.028
Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform
Javier Fernandez-Martinez et al. Cell. 2016.
Abstract
The last steps in mRNA export and remodeling are performed by the Nup82 complex, a large conserved assembly at the cytoplasmic face of the nuclear pore complex (NPC). By integrating diverse structural data, we have determined the molecular architecture of the native Nup82 complex at subnanometer precision. The complex consists of two compositionally identical multiprotein subunits that adopt different configurations. The Nup82 complex fits into the NPC through the outer ring Nup84 complex. Our map shows that this entire 14-MDa Nup82-Nup84 complex assembly positions the cytoplasmic mRNA export factor docking sites and messenger ribonucleoprotein (mRNP) remodeling machinery right over the NPC's central channel rather than on distal cytoplasmic filaments, as previously supposed. We suggest that this configuration efficiently captures and remodels exporting mRNP particles immediately upon reaching the cytoplasmic side of the NPC.
Keywords: Nup4 complex; Nup82 complex; computational structural biology; cross-linking and mass spectrometry; electron microscopy; integrative structure determination; mRNA export; mRNP remodeling; nuclear pore complex; small-angle X-ray scattering.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
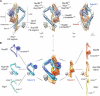
Figure 1. Structure of the core Nup82 holo-complex
(A) Three views of the localization probability density map corresponding to the Nup82 holo-complex ensemble are shown (light gray), with a single representative ribbon structure embedded; the proteins, subunits, and different structural features of the complex are indicated. Subunit assignment is indicated with a superscript “s1” (subunit 1) or “s2” (subunit 2). In all views the components of each subunit are colored in tones of red (subunit 1) or blue (subunit 2) (see also panel B). (B) Exploded view of the Nup82 holo-complex subunits and protein components, with the whole complex shown in the center and the two subunits and the different components shown on the right (subunit 1, colored in red tones) or the left side (subunit 2, colored in blue tones). CCS indicates “Coiled Coil Segment” as described in the main text. See also Figures S1, S2, S3 and Tables S1 and S2.
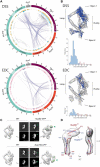
Figure 2. Nup82 holo-complex structure validation
(A) Circos-XL plots showing the distribution of all DSS (upper plot) or EDC (lower plot) cross-links mapping within the core of the Nup82 holo-complex. Each protein is represented as a colored segment, with the amino acid residue indicated on the outside of the plot and relevant domains indicated inside each segment; regions without reliable fold assignment are identified by lighter shading. Inter-molecular cross-links are depicted as purple lines and intra-molecular cross-links as gray lines. The internal circles include bars representing the density of cross-links per 10 residues in DSS and EDC (blue and light blue color for inter-molecular cross-links and intra-molecular cross-links, respectively) and the density of lysines in DSS (orange and light orange bars for cross-linked and uncross-linked residues, respectively) or the density of lysine/carboxylic acid in EDC (pink and light pink bars for cross-linked and uncross-linked residues, respectively). (B) Structure of the Nup82 holo-complex showing the cross-links falling within the expected Cα–Cα maximum distance threshold (blue) or outside of that threshold (orange). Below the structure, a bar graph shows the Cα–Cα distance distribution of all DSS or EDC cross-links in the structure. DSS threshold = 35 Å and EDC threshold = 30 Å. (C) GFP mass-tagging analysis of the Nup82 holo-complex: Analyses of a Nup82-GFP tagged version (upper diagram) or a Nup159-GFP tagged version (lower diagram) of the holo-complex are shown. For each diagram, a view of the native Nup82 holocomplex structure is shown (WT), and the tagged version of the structure shown on the right side. The upper panels show a representative negative stain 2D class average of the native complex (left panel) and the tagged version (right panel; green arrowhead, GFP). The lower panels show 2D projections of the native structure (left panel) and the calculated GFP-tagged version (right panel; green arrowhead, GFP). ccc, cross correlation coefficient; Scale bar, 10 nm. (D) SAXS analysis of the Nup82(572-690) fragment, showing two views of the computed ab initio shape (gray envelope), with ribbon representations of the equivalent Nup82 fragments in the conformation they adopt within the Nup82 holo-complex; subunits 1 (red) and 2 (blue) are indicated. See also Figure S5D-F and Table S4.
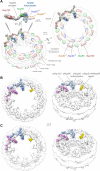
Figure 3. Molecular architecture of the cytoplasmic mRNA export and remodeling platform
(A) Structure of the Nup82-Nup84 complex assembly. Three views of the structural arrangement formed by the Y-shaped Nup84 complex (light gray density) and the Nup82 holo-complex (light blue density) calculated using CX-MS data. Each component and structural feature of the different complexes are labeled and shown as a density with fitted ribbon representations of their component nups. A circos plot shows the distribution of cross-links (dashed, light blue lines) identified between components of the Nup82 complex, Nup84 complex, and mRNP export/remodeling machinery, used for the calculation of the assembly and the map described in panel B. (B) Molecular architecture of the cytoplasmic mRNA export and remodeling platform. An exploded view of the the different platform components is presented (solid blue lines, covalent attachment; dashed blue lines, CX-MS identified associations). When available, components are represented as crystal structures (Dbp5, Gle1 and Nup159 N-termini, pdb: 3RRM (Montpetit et al., 2011); Gle2/RAE1, pdb: 3MMY (Ren et al., 2010); Nup116 C-termini, pdb: 3PBP (Yoshida et al., 2011) and 3NF5 (Sampathkumar et al., 2012)). The Gle1 N-terminus is represented with a homology model of its predicted coiled-coil region inside a light red density of the approximate expected size for the domain. See also Table S3.
Figure 4. mRNA export phenotype in Nup84 complex mutants is associated to defective incorporation of the Nup82 holo-complex into the NPC
(A) The mRNA export defect phenotype was quantified and plotted (mean value; n = 4) for each Nup84 complex component mutant in order of increasing level of nuclear polyA mRNA accumulation as observed by FISH (see STAR Methods and Figure S6 for details), and assigned to five divisions of increasing level of accumulation (white to dark purple)(Fernandez-Martinez et al., 2012). Representative examples of strains included in each division are shown on the top. AU, arbitrary unit; Errors = SEM; Scale bar, 5 microns. (B) Mapping of the color code described in (A) into the Nup84 complex components. Horizontal lines represent the amino acid residue length of each protein and truncated version; amino acid residue positions are shown on top of the lines. (C) The severity of nuclear mRNA accumulation phenotype (detailed in panels A and B) for specific truncations of the Nup84 complex components are shown mapped into the Nup82-Nup84 complex assembly. The color code is the same as the one described in panel A. The Nup82 holo-complex density is shown in light blue. (D) Subcellular localization of Nup82-GFP in Nup84 complex truncation mutants. Top, diagrams representing the Nup84 complex, with the corresponding truncated region of the complex shown. Middle panels show the localization of the genomically tagged Nup82-GFP reporter as determined by fluorescence microscopy. Lower panels show the differential interference contrast (DIC) image of the same cells. Scale bar, 5 microns. See also Figure S6.
Figure 5. Position of the Nup82-Nup84 complex assembly within the NPC
(A) Fitting to the yeast NPC map. Two views of the optimized alignment of two_S. cerevisiae_ Nup82-Nup84 complex assemblies into the_S. cerevisiae_ NPC localization probability density map (transparent gray), together with a side view of the detailed alignment (Alber et al., 2007b); Nup85 (green), Nup133 (red), and two Nup82 units (blue and orange) are indicated. Scale bars, 100 Å. (B) Comparison with the human NPC tomographic cryo-EM map (EMDB 2444) (Bui et al., 2013). Two views of the optimized alignment of two S. cerevisiae Nup82-Nup84 complex assemblies (pink and blue) into the human NPC map (CCC = 0.72). One suggested localization for the human Nup214/Nup88 complex is colored in yellow. (C) Comparison with the mutant human NPC tomographic cryo-EM map (EMDB 3104) (von Appen et al., 2015), lacking an outer cytoplasmic Y-complex ring (CCC = 0.81). See also Figure S7.
Figure 6. The Nup82-Nup84 complex assembly acts as a scaffold to organize the FG region conveyor belts and mRNP remodeling sites in the NPC
Upper: model for the arrangement of the FG regions associated to the Nup82 holo-complex. FG regions were modeled using molecular dynamics. The position of the Nup116 FG regions is based on the position of their C-termini (PDB: 3PBP (Yoshida et al., 2011), but could vary significantly, depending on the orientation of the unstructured region connecting the FG domains (dotted blue line). N-termini of Nup159 can interact during mRNP remodeling with Dbp5 as indicated by dashed blue line. Sequential mRNP export and remodeling steps associated with each region of the complex are shown on the left. Lower left: mapping of disease-associated Gle1 mutations into our model for the mRNA export platform. The yeast Gle1 region equivalent to where disease-related mutations have been found in human Gle1 were colored in purple (lethal congenital contracture syndrome 1 (LCCS-1)), gold (lethal arthrogryposis with anterior horn cell disease (LAAHD)) and cyan (amyotrophic lateral sclerosis (ALS)), based on data described in (Folkmann et al., 2014; Kaneb et al., 2015; Kendirgi et al., 2003). Proteins are represented as described in Figure 3B. Dashed blue lines indicate identified protein-protein associations. Lower right: schematic representation comparing the previous view (left) of the Nup82 complex as components of cytoplasmically oriented filaments, with the new view (right) of how it instead forms struts projecting towards the NPC central channel, positions the FG regions to fill the channel and forms the top part of the central transporter region. See also Table S3.
Comment in
- A New Path through the Nuclear Pore.
Gozalo A, Capelson M. Gozalo A, et al. Cell. 2016 Nov 17;167(5):1159-1160. doi: 10.1016/j.cell.2016.11.011. Cell. 2016. PMID: 27863235
Similar articles
- Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold.
Gaik M, Flemming D, von Appen A, Kastritis P, Mücke N, Fischer J, Stelter P, Ori A, Bui KH, Baßler J, Barbar E, Beck M, Hurt E. Gaik M, et al. J Cell Biol. 2015 Feb 2;208(3):283-97. doi: 10.1083/jcb.201411003. J Cell Biol. 2015. PMID: 25646085 Free PMC article. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Into the basket and beyond: the journey of mRNA through the nuclear pore complex.
Ashkenazy-Titelman A, Shav-Tal Y, Kehlenbach RH. Ashkenazy-Titelman A, et al. Biochem J. 2020 Jan 17;477(1):23-44. doi: 10.1042/BCJ20190132. Biochem J. 2020. PMID: 31913454 Review. - Single particle imaging of mRNAs crossing the nuclear pore: Surfing on the edge.
Palazzo AF, Truong M. Palazzo AF, et al. Bioessays. 2016 Aug;38(8):744-50. doi: 10.1002/bies.201600038. Epub 2016 Jun 8. Bioessays. 2016. PMID: 27276446 Review.
Cited by
- Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Jishage M, Yu X, Shi Y, Ganesan SJ, Chen WY, Sali A, Chait BT, Asturias FJ, Roeder RG. Jishage M, et al. Nat Struct Mol Biol. 2018 Sep;25(9):859-867. doi: 10.1038/s41594-018-0118-5. Epub 2018 Sep 6. Nat Struct Mol Biol. 2018. PMID: 30190596 Free PMC article. - DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes.
Meinema AC, Marzelliusardottir A, Mirkovic M, Aspert T, Lee SS, Charvin G, Barral Y. Meinema AC, et al. Elife. 2022 Apr 4;11:e71196. doi: 10.7554/eLife.71196. Elife. 2022. PMID: 35373738 Free PMC article. - Archiving and disseminating integrative structure models.
Vallat B, Webb B, Westbrook J, Sali A, Berman HM. Vallat B, et al. J Biomol NMR. 2019 Jul;73(6-7):385-398. doi: 10.1007/s10858-019-00264-2. Epub 2019 Jul 5. J Biomol NMR. 2019. PMID: 31278630 Free PMC article. - The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress.
Wang X, Chemmama IE, Yu C, Huszagh A, Xu Y, Viner R, Block SA, Cimermancic P, Rychnovsky SD, Ye Y, Sali A, Huang L. Wang X, et al. J Biol Chem. 2017 Sep 29;292(39):16310-16320. doi: 10.1074/jbc.M117.803619. Epub 2017 Aug 15. J Biol Chem. 2017. PMID: 28821611 Free PMC article. - Mechanisms of nuclear mRNA export: A structural perspective.
Xie Y, Ren Y. Xie Y, et al. Traffic. 2019 Nov;20(11):829-840. doi: 10.1111/tra.12691. Epub 2019 Sep 12. Traffic. 2019. PMID: 31513326 Free PMC article. Review.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS yeast research. 2012;12:88–96. - PubMed
- Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008284/GM/NIGMS NIH HHS/United States
- R01 GM083960/GM/NIGMS NIH HHS/United States
- P41 GM103314/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- R01 GM112108/GM/NIGMS NIH HHS/United States
- CIHR/Canada
- U54 GM103511/GM/NIGMS NIH HHS/United States
- P41 GM109824/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials