A DNA methylation biomarker of alcohol consumption - PubMed (original) (raw)
Meta-Analysis
. 2018 Feb;23(2):422-433.
doi: 10.1038/mp.2016.192. Epub 2016 Nov 15.
C Liu 1 2 3, R E Marioni 4 5 6, L Pfeiffer 8 9, P-C Tsai 10, L M Reynolds 11, A C Just 12, Q Duan 13, C G Boer 14, T Tanaka 15, C E Elks 16, S Aslibekyan 17, J A Brody 18, B Kühnel 8 9, C Herder 19 20, L M Almli 21, D Zhi 22, Y Wang 23, T Huan 1 2, C Yao 1 2, M M Mendelson 1 2, R Joehanes 1 2 24, L Liang 25, S-A Love 23, W Guan 26, S Shah 6 27, A F McRae 6 27, A Kretschmer 8 9, H Prokisch 28 29, K Strauch 30 31, A Peters 8 9 32, P M Visscher 4 6 27, N R Wray 6 27, X Guo 33, K L Wiggins 18, A K Smith 21, E B Binder 34, K J Ressler 35, M R Irvin 17, D M Absher 36, D Hernandez 37, L Ferrucci 15, S Bandinelli 38, K Lohman 11, J Ding 39, L Trevisi 40, S Gustafsson 7, J H Sandling 41 42, L Stolk 14, A G Uitterlinden 14 43, I Yet 10, J E Castillo-Fernandez 10, T D Spector 10, J D Schwartz 44, P Vokonas 45, L Lind 46, Y Li 47, M Fornage 48, D K Arnett 49, N J Wareham 16, N Sotoodehnia 18, K K Ong 16, J B J van Meurs 14, K N Conneely 50, A A Baccarelli 51, I J Deary 4 52, J T Bell 10, K E North 23, Y Liu 11, M Waldenberger 8 9, S J London 53, E Ingelsson 7 54, D Levy 1 2
Affiliations
- PMID: 27843151
- PMCID: PMC5575985
- DOI: 10.1038/mp.2016.192
Meta-Analysis
A DNA methylation biomarker of alcohol consumption
C Liu et al. Mol Psychiatry. 2018 Feb.
Abstract
The lack of reliable measures of alcohol intake is a major obstacle to the diagnosis and treatment of alcohol-related diseases. Epigenetic modifications such as DNA methylation may provide novel biomarkers of alcohol use. To examine this possibility, we performed an epigenome-wide association study of methylation of cytosine-phosphate-guanine dinucleotide (CpG) sites in relation to alcohol intake in 13 population-based cohorts (ntotal=13 317; 54% women; mean age across cohorts 42-76 years) using whole blood (9643 European and 2423 African ancestries) or monocyte-derived DNA (588 European, 263 African and 400 Hispanic ancestry) samples. We performed meta-analysis and variable selection in whole-blood samples of people of European ancestry (n=6926) and identified 144 CpGs that provided substantial discrimination (area under the curve=0.90-0.99) for current heavy alcohol intake (⩾42 g per day in men and ⩾28 g per day in women) in four replication cohorts. The ancestry-stratified meta-analysis in whole blood identified 328 (9643 European ancestry samples) and 165 (2423 African ancestry samples) alcohol-related CpGs at Bonferroni-adjusted P<1 × 10-7. Analysis of the monocyte-derived DNA (n=1251) identified 62 alcohol-related CpGs at P<1 × 10-7. In whole-blood samples of people of European ancestry, we detected differential methylation in two neurotransmitter receptor genes, the γ-Aminobutyric acid-A receptor delta and γ-aminobutyric acid B receptor subunit 1; their differential methylation was associated with expression levels of a number of genes involved in immune function. In conclusion, we have identified a robust alcohol-related DNA methylation signature and shown the potential utility of DNA methylation as a clinically useful diagnostic test to detect current heavy alcohol consumption.
Conflict of interest statement
Erik Ingelsson is an advisor and consultant for Precision Wellness Inc (Redwood City, CA). The remaining authors declare no conflict of interest.
Figures
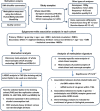
Figure 1
Overview of the study design. ARIC, The Atherosclerosis Risk in Communities study; BMI, body mass index; DNAm, DNA methylation value; FHS, the Framingham Heart Study; I (light/at-risk/heavy drinkers versus non-drinkers), the indicator variable for light drinkers versus non-drinkers, at-risk drinkers versus non-drinkers and heavy drinkers versus non-drinkers; KORA F4, The Cooperative Health Research in the Region of Augsburg study; LASSO, least absolute shrinkage and selection operator; LBC, The Lothian Birth Cohort; MESA, The Multi-Ethnic Study of Atherosclerosis; WBCs, white blood cell counts.
Figure 2
A biomarker of heavy alcohol drinking. Four sets of cytosine-phosphate-guanine dinucleotides (CpGs) were selected at _s_=0.12 (5 CpGs), _s_=0.08 (23 CpGs), _s_=‘lambda.1se’ (78 CpGs) and _s_=‘lambda.min’ (144 CpGs) using least absolute shrinkage and selection operator (LASSO) in the Framingham Heart Study (FHS) cohort (the training cohort). ROC analysis was performed to classify heavy drinkers versus non-drinkers (left figure) and heavy drinkers versus light drinkers (right figure). ‘Non-drinkers’ were subjects with no alcohol consumption (i.e., g per day=0); ‘light drinkers’ were subjects who consumed 0<g per day⩽28 in men and 0<g per day⩽14 in women; ‘heavy drinkers’ were subjects who consumed ⩾42 g per day in men and ⩾28 g per day in women. ARIC, The Atherosclerosis Risk in Communities study; KORA F4, The Cooperative Health Research in the Region of Augsburg study; LBC1936, The Lothian Birth Cohort 1936; MESA, The Multi-Ethnic Study of Atherosclerosis.
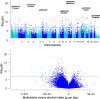
Figure 3
Meta-analysis of epigenome-wide association of alcohol intake in European ancestry (EA) whole-blood samples: the Manhattan plot (top) and the volcano plot (bottom). The DNA methylation proportion was the outcome variable, grams alcohol consumed per day (g per day) was the predictor variable, adjusting for age, sex, body mass index, technical covariates and white blood cell counts. The inverse-variance weighted random-effects model was performed in meta-analysis using all whole blood DNA samples of EA.
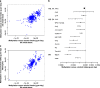
Figure 4
Comparison of regression coefficients of the significant cytosine-phosphate-guanine dinucleotides (CpGs) in association analysis of the continuous alcohol trait (g per day): (a) between European and African whole-blood samples; (b) the Forest plot of effect estimates and standard errors of cg11376147 in all study cohorts; and (c) between European whole-blood and CD14+ monocyte samples. (a) Includes a list of CpGs with P<1 × 10−7 in EA or AA whole-blood samples and (c) includes a list of CpGs with P<1 × 10−7 in EA whole-blood samples or in monocyte samples of mixed ancestries. The Pearson’s correlation was _r_=0.64 between the effect estimates in (a) and _r_=0.72 in (c). MM, monocyte, mixed ancestries; WB AA, whole blood, African ancestry; WB EA, whole blood, European ancestry.
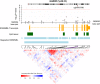
Figure 5
The γ-aminobutyric acid-A (GABA-A) receptor, delta (GABRD): the associations of the 36 cytosine-phosphate-guanine dinucleotides (CpGs) within GABRD, genomic and regulatory features and correlation of methylation measurements. The results were obtained in meta-analysis of the association analysis of 9643 whole-blood-derived DNA samples of European ancestry (EA) individuals. The correlation of these 36 CpGs was calculated using the methylation measurements at 36 CpGs, adjusting for age, sex, technical covariates and white cell blood counts in the Framingham Heart Study samples.
Comment in
- Overestimation of the classification accuracy of a biomarker for assessing heavy alcohol use.
Hattab MW, Clark SL, van den Oord EJCG. Hattab MW, et al. Mol Psychiatry. 2018 Nov;23(11):2114-2115. doi: 10.1038/mp.2017.181. Mol Psychiatry. 2018. PMID: 28894303 Free PMC article. No abstract available.
Similar articles
- Epigenome-Wide DNA Methylation Association Analysis Identified Novel Loci in Peripheral Cells for Alcohol Consumption Among European American Male Veterans.
Xu K, Montalvo-Ortiz JL, Zhang X, Southwick SM, Krystal JH, Pietrzak RH, Gelernter J. Xu K, et al. Alcohol Clin Exp Res. 2019 Oct;43(10):2111-2121. doi: 10.1111/acer.14168. Epub 2019 Aug 30. Alcohol Clin Exp Res. 2019. PMID: 31386212 Free PMC article. - Blood-based epigenome-wide analyses of 19 common disease states: A longitudinal, population-based linked cohort study of 18,413 Scottish individuals.
Hillary RF, McCartney DL, Smith HM, Bernabeu E, Gadd DA, Chybowska AD, Cheng Y, Murphy L, Wrobel N, Campbell A, Walker RM, Hayward C, Evans KL, McIntosh AM, Marioni RE. Hillary RF, et al. PLoS Med. 2023 Jul 6;20(7):e1004247. doi: 10.1371/journal.pmed.1004247. eCollection 2023 Jul. PLoS Med. 2023. PMID: 37410739 Free PMC article. Review. - Alcohol consumption is associated with widespread changes in blood DNA methylation: Analysis of cross-sectional and longitudinal data.
Dugué PA, Wilson R, Lehne B, Jayasekara H, Wang X, Jung CH, Joo JE, Makalic E, Schmidt DF, Baglietto L, Severi G, Gieger C, Ladwig KH, Peters A, Kooner JS, Southey MC, English DR, Waldenberger M, Chambers JC, Giles GG, Milne RL. Dugué PA, et al. Addict Biol. 2021 Jan;26(1):e12855. doi: 10.1111/adb.12855. Epub 2019 Dec 2. Addict Biol. 2021. PMID: 31789449 - Whole Blood DNA Methylation Signatures of Diet Are Associated With Cardiovascular Disease Risk Factors and All-Cause Mortality.
Ma J, Rebholz CM, Braun KVE, Reynolds LM, Aslibekyan S, Xia R, Biligowda NG, Huan T, Liu C, Mendelson MM, Joehanes R, Hu EA, Vitolins MZ, Wood AC, Lohman K, Ochoa-Rosales C, van Meurs J, Uitterlinden A, Liu Y, Elhadad MA, Heier M, Waldenberger M, Peters A, Colicino E, Whitsel EA, Baldassari A, Gharib SA, Sotoodehnia N, Brody JA, Sitlani CM, Tanaka T, Hill WD, Corley J, Deary IJ, Zhang Y, Schöttker B, Brenner H, Walker ME, Ye S, Nguyen S, Pankow J, Demerath EW, Zheng Y, Hou L, Liang L, Lichtenstein AH, Hu FB, Fornage M, Voortman T, Levy D. Ma J, et al. Circ Genom Precis Med. 2020 Aug;13(4):e002766. doi: 10.1161/CIRCGEN.119.002766. Epub 2020 Jun 11. Circ Genom Precis Med. 2020. PMID: 32525743 Free PMC article. - DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study.
Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H, Wolffenbuttel BHR, van Vliet-Ostaptchouk JV. Walaszczyk E, et al. Diabetologia. 2018 Feb;61(2):354-368. doi: 10.1007/s00125-017-4497-7. Epub 2017 Nov 21. Diabetologia. 2018. PMID: 29164275 Free PMC article. Review.
Cited by
- Association analysis between an epigenetic alcohol risk score and blood pressure.
Bui H, Keshawarz A, Wang M, Lee M, Ratliff SM, Lin L, Birditt KS, Faul JD, Peters A, Gieger C, Delerue T, Kardia SLR, Zhao W, Guo X, Yao J, Rotter JI, Li Y, Liu X, Liu D, Tavares JF, Pehlivan G, Breteler MMB, Karabegovic I, Ochoa-Rosales C, Voortman T, Ghanbari M, van Meurs JBJ, Nasr MK, Dörr M, Grabe HJ, London SJ, Teumer A, Waldenberger M, Weir DR, Smith JA, Levy D, Ma J, Liu C. Bui H, et al. Clin Epigenetics. 2024 Oct 28;16(1):149. doi: 10.1186/s13148-024-01753-4. Clin Epigenetics. 2024. PMID: 39468603 Free PMC article. - Association of smoking habits with TXNIP DNA methylation levels in leukocytes among general Japanese population.
Maeda K, Yamada H, Munetsuna E, Fujii R, Yamazaki M, Ando Y, Mizuno G, Ishikawa H, Ohashi K, Tsuboi Y, Hashimoto S, Hamajima N, Suzuki K. Maeda K, et al. PLoS One. 2020 Jul 1;15(7):e0235486. doi: 10.1371/journal.pone.0235486. eCollection 2020. PLoS One. 2020. PMID: 32609762 Free PMC article. - The World of Oral Cancer and Its Risk Factors Viewed from the Aspect of MicroRNA Expression Patterns.
Aghiorghiesei O, Zanoaga O, Nutu A, Braicu C, Campian RS, Lucaciu O, Berindan Neagoe I. Aghiorghiesei O, et al. Genes (Basel). 2022 Mar 26;13(4):594. doi: 10.3390/genes13040594. Genes (Basel). 2022. PMID: 35456400 Free PMC article. Review. - Association of leukocyte DNA methylation changes with dietary folate and alcohol intake in the EPIC study.
Perrier F, Viallon V, Ambatipudi S, Ghantous A, Cuenin C, Hernandez-Vargas H, Chajès V, Baglietto L, Matejcic M, Moreno-Macias H, Kühn T, Boeing H, Karakatsani A, Kotanidou A, Trichopoulou A, Sieri S, Panico S, Fasanelli F, Dolle M, Onland-Moret C, Sluijs I, Weiderpass E, Quirós JR, Agudo A, Huerta JM, Ardanaz E, Dorronsoro M, Tong TYN, Tsilidis K, Riboli E, Gunter MJ, Herceg Z, Ferrari P, Romieu I. Perrier F, et al. Clin Epigenetics. 2019 Apr 2;11(1):57. doi: 10.1186/s13148-019-0637-x. Clin Epigenetics. 2019. PMID: 30940212 Free PMC article. - Epigenetic prediction of complex traits and mortality in a cohort of individuals with oropharyngeal cancer.
Langdon RJ, Beynon RA, Ingarfield K, Marioni RE, McCartney DL, Martin RM, Ness AR, Pawlita M, Waterboer T, Relton C, Thomas SJ, Richmond RC. Langdon RJ, et al. Clin Epigenetics. 2020 Apr 22;12(1):58. doi: 10.1186/s13148-020-00850-4. Clin Epigenetics. 2020. PMID: 32321578 Free PMC article.
References
- NIAAA. Alcohol Facts and Statistics. Available at: https://www.niaaa.nih.gov/alcohol-health/overview-alcohol-consumption/al....
- Ogeil RP, Room R, Matthews S, Lloyd B. Alcohol and burden of disease in Australia: the challenge in assessing consumption. Aust NZ J Public Health 2015; 39: 121–123. - PubMed
- Rehm J, Taylor B, Roerecke M, Patra J. Alcohol consumption and alcohol-attributable burden of disease in Switzerland, 2002. Int J Public Health 2007; 52: 383–392. - PubMed
- Ferreira-Borges C, Rehm J, Dias S, Babor T, Parry CD. The impact of alcohol consumption on African people in 2012: an analysis of burden of disease. Trop Med Int Health 2015; 21: 52–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 ES015172/ES/NIEHS NIH HHS/United States
- N01HC95159/HL/NHLBI NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- N01HC95164/HL/NHLBI NIH HHS/United States
- 14136/CRUK_/Cancer Research UK/United Kingdom
- G0701120/MRC_/Medical Research Council/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- N01HC85080/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- N01HC95169/HL/NHLBI NIH HHS/United States
- R01 HG006292/HG/NHGRI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- N01HC95161/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- R01 HL129132/HL/NHLBI NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- G0401527/MRC_/Medical Research Council/United Kingdom
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- N01HC95168/HL/NHLBI NIH HHS/United States
- MC_UU_12015/1/MRC_/Medical Research Council/United Kingdom
- C864/A8257/CRUK_/Cancer Research UK/United Kingdom
- R01 HL092111/HL/NHLBI NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 HL101250/HL/NHLBI NIH HHS/United States
- MR/N003284/1/MRC_/Medical Research Council/United Kingdom
- N01HC95162/HL/NHLBI NIH HHS/United States
- MR/M013111/1/MRC_/Medical Research Council/United Kingdom
- G9502233/MRC_/Medical Research Council/United Kingdom
- N01HC85082/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 ES021733/ES/NIEHS NIH HHS/United States
- N01HC95165/HL/NHLBI NIH HHS/United States
- G1001245/MRC_/Medical Research Council/United Kingdom
- MC_UU_12015/2/MRC_/Medical Research Council/United Kingdom
- N01HC55222/HL/NHLBI NIH HHS/United States
- R01 HL111089/HL/NHLBI NIH HHS/United States
- UL1 TR000454/TR/NCATS NIH HHS/United States
- N01HC85079/HL/NHLBI NIH HHS/United States
- N01HC95167/HL/NHLBI NIH HHS/United States
- N01HC85083/HL/NHLBI NIH HHS/United States
- N01HC95163/HL/NHLBI NIH HHS/United States
- N01HC85086/HL/NHLBI NIH HHS/United States
- MR/L00002/1/MRC_/Medical Research Council/United Kingdom
- G1000143/MRC_/Medical Research Council/United Kingdom
- N01HC25195/HL/NHLBI NIH HHS/United States
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- R21 AA027450/AA/NIAAA NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- Z99 HL999999/ImNIH/Intramural NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- N01HC95166/HL/NHLBI NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- R01 HL116747/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01HC85081/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- N01HC95160/HL/NHLBI NIH HHS/United States
- K99 HL136875/HL/NHLBI NIH HHS/United States
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical