β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance - PubMed (original) (raw)
. 2016 Nov 17;167(5):1354-1368.e14.
doi: 10.1016/j.cell.2016.09.034.
Ehsan Habibi 1, Shuang-Yin Wang 1, Rob J W Arts 2, Robab Davar 1, Wout Megchelenbrink 1, Bowon Kim 1, Tatyana Kuznetsova 1, Matthijs Kox 3, Jelle Zwaag 3, Filomena Matarese 1, Simon J van Heeringen 4, Eva M Janssen-Megens 1, Nilofar Sharifi 1, Cheng Wang 1, Farid Keramati 1, Vivien Schoonenberg 1, Paul Flicek 5, Laura Clarke 5, Peter Pickkers 3, Simon Heath 6, Ivo Gut 6, Mihai G Netea 2, Joost H A Martens 1, Colin Logie 1, Hendrik G Stunnenberg 7
Affiliations
- PMID: 27863248
- PMCID: PMC5927328
- DOI: 10.1016/j.cell.2016.09.034
β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance
Boris Novakovic et al. Cell. 2016.
Abstract
Innate immune memory is the phenomenon whereby innate immune cells such as monocytes or macrophages undergo functional reprogramming after exposure to microbial components such as lipopolysaccharide (LPS). We apply an integrated epigenomic approach to characterize the molecular events involved in LPS-induced tolerance in a time-dependent manner. Mechanistically, LPS-treated monocytes fail to accumulate active histone marks at promoter and enhancers of genes in the lipid metabolism and phagocytic pathways. Transcriptional inactivity in response to a second LPS exposure in tolerized macrophages is accompanied by failure to deposit active histone marks at promoters of tolerized genes. In contrast, β-glucan partially reverses the LPS-induced tolerance in vitro. Importantly, ex vivo β-glucan treatment of monocytes from volunteers with experimental endotoxemia re-instates their capacity for cytokine production. Tolerance is reversed at the level of distal element histone modification and transcriptional reactivation of otherwise unresponsive genes. VIDEO ABSTRACT.
Keywords: BLUEPRINT; endotoxin tolerance; epigenome; histone modifications; human monocytes; lipopolysaccharide; macrophages; trained innate immunity; transcriptome; β-glucan.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Figure 1. Epigenomic and Transcriptomic Remodeling of Monocytes Induced by Exposure to LPS or BG
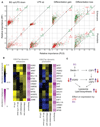
(A) Experimental setup for epigenomic interrogation of monocyte-to-macrophage differentiation and induction of tolerance (with LPS) or trained immunity (with BG). (B) PCA plots of H3K27ac and H3K4me1 dynamic enhancers (monocytes, red circle; naive, circle; LPS, triangle; BG, square; 1 hr, blue; 4 hr, black; day 1, green; and day 6, brown). Dynamic H3K27ac patterns show a clear deviation from the differentiation pathway (PC1) in LPS-treated cells. On the other hand, BG-treated cells at day 1 are well on their way toward a full macrophage epigenetic profile. (C) A total of 17,500 H3K27ac dynamic gene-distal regions were identified and can be clearly separated into four clusters: BG up/LPS down, LPS up, differentiation gain, and differentiation loss. Solid lines are median log-FC relative to day 0, and shaded areas represent the 25th and 75th quartile. Naive cells are shown as a green line, LPS as a red line, and BG as a purple line. H3K4me1 at these regions can be seen in Figure S1B; LPS induces early H3K27ac accumulation, followed by long-term H3K4me1 marking, while BG induces concurrent accumulation of H3K27ac and H3K4me1. (D) PCA plots showing the relationships among all samples based on dynamic gene expression. PC1 explains most of the variation and is associated with differentiation. PC2 is LPS related, with LPS 4 hr and LPS day 1 samples separating from the corresponding naive and BG samples. (E) Heatmap of differentiation associated genes, as well as those induced by LPS or BG exposure. The general trend in expression is that BG exposed cells start to express differentiation associated genes faster (at day 1) than naive cells, while LPS exposed cells lag behind. (F) Top pathways associated with differentiation and showing opposing directions in response to BG and LPS. See also Figures S1, S2, and S3 and Tables S1, S2, S3, and S4.
Figure 2. Motif Enrichment at Epigenetically Dynamic Promoters and Enhancers and Associated Transcription Factor Networks
Motif enrichment analysis was performed on ATAC-sequencing (nucleosome-free) peaks that overlap H3K27ac dynamic enhancers and H3K27ac promoters. (A) Random forest (RF) and a partial least-squares (PLS) classifiers were trained using the TF motifs found by GIMME to determine features (TF motifs) based on their ability to separate the 4 H3K27ac clusters shown in Figure 1C. Both classifiers produce a feature importance score (between 0 and 100), which is a measure of how “characteristic” the presence or absence of the TF motif is for the considered cluster. Green dots represent positive features (motif over-represented in cluster), and red dots represent negative features (motif under-represented in cluster). The EGR2 motif was the strongest positive feature for the BG up/LPS down enhancer cluster, NF-κB for the LPS up cluster, SPI1 (PU.1) for the differentiation gain cluster, and JUNB for the differentiation loss cluster. At the promoter regions NF-κB was a positive feature for LPS up cluster, MITF for the differentiation gain cluster, and CREB1 and JUNB for the differentiation loss cluster. (B) Motif enrichment is plotted as absolute difference in abundance compared to background (yellow, higher abundance than background; blue, lower abundance than background) for the top enriched motifs. Consistently identified transcription factor motifs include SPI1 at differentiation associated enhancers, NF-κB at LPS enhancers, and EGR2 and MITF at BG enhancers. Abundance increase over background supports the level of importance score. (C) A diagram of the transcription factor network based on EGR2 and MITF motif occurrence at BG-induced lysosome and lipid metabolism genes. Purple arrows indicate the direction of expression induced by BG exposure, and red arrows indicate the direction of expression induced by for LPS exposure. BG exposure induces transient expression of the genes, while LPS exposure inhibits activation. The full network based on promoter abundance is shown in Figure S4B. See also Figure S4.
Figure 3. Macrophage Endotoxin Tolerance Defined at the Transcriptional Level following LPS Re-exposure
(A) The innate immune memory model, including data collection at LPS re-exposure at day 6. (B) PCA plots of dynamic RNA-seq, H3K27ac at promoters and enhancers, and H3K4me1 peaks, including LPS re-exposure samples. After re-exposure to LPS, significant enhancer H3K27ac changes occur in LPS-Mfs, indicating that they are capable of activating their enhancers. However, the level of their response is lower compared to monocytes, naive-Mfs, and BG-Mfs, which can be seen on the second principal component. Unlike RNA and H3K27ac, H3K4me1 does not show significant changes following LPS re-exposure in any of the three macrophage subtypes. (C) The total macrophage transcriptional response (750 genes) to LPS was separated into three groups based on the induction of genes in LPS-Mfs, relative to naive-Mfs and BG-Mfs, revealing a gradient in LPS-Mf response to LPS re-exposure. The groups are (G1) tolerized genes, (G2) partially tolerized genes, and (G3) responsive genes. See also Figure S5.
Figure 4. Histone Modification Dynamics and Open Chromatin Analysis at Tolerized Gene Promoters
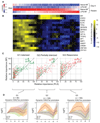
(A) Heatmap showing average expression of 777 LPS-responsive genes in naive-Mfs. Genes are ranked based on their induction in LPS-Mfs, first by tolerance group (G1, G2, and G3) and then by relative induction compared to naive-Mfs within each group. Response to LPS re-exposure is a gradient in LPS-Mfs, with the most tolerized genes on the left and the most responsive genes on the right. (B) Heatmap showing abundance of significant motifs in the promoter regions of the three macrophage LPS-responsive gene groups. The tolerized gene promoters are enriched for several transcriptional repressors, such as EGR2 and TP53, while the partially tolerized gene promoters are enriched for IRF and STAT motifs. (C) Random forest (RF) and a partial least-squares (PLS) classifiers importance score (between 0 and 100) for each tolerized gene cluster (G1, tolerized; G2, partially tolerized; and G3, responsive). Green dots represent over-represented motifs, and red dots under-represented motifs. The top features of G1 gene promoters are E2F3, EGR2, and ZBTB7B motifs. The top features for G2 gene promoters are IRF and STAT, while G3 promoters do not have over-represented features but are depleted of EGR2, E2F3, and ZNF350. (D) Median H3K27ac at dynamic promoters of G1, G2, and G3 group genes, shaded areas represent the 25th and 75th quartile. This shows that LPS-Mfs do not accumulate H3K27ac at tolerized genes but do so at the promoters of responsive genes. See also Figure S6 for H3K4me3. See also Figure S6.
Figure 5. BG Can Reverse Both In Vitro and In Vivo LPS-Induced Tolerance and Reinstate Proper Cytokine Production in Macrophages
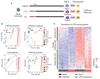
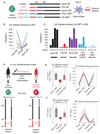
(A) The in vitro monocyte tolerance reversal model, with BG added therapeutically after 24 hr of LPS exposure (rescue-Mfs). The histone-mimic and inflammation blocker IBET was used in a preventative (co-culture with LPS for 24 hr LPS-co-IBET-Mfs) and a therapeutic (added after 24 hr of LPS exposure [LPS + IBET-Mfs]) manner. Following several days of rest, macrophages were re-exposed to LPS and cytokine release measured after 24 hr. (B) BG re-instates IL-6 release in tolerized macrophages. Data from six donors are shown for naive-Mfs, LPS-Mfs, and rescue-Mfs. (C) Preventative use of IBET blocks the first LPS response in monocytes, resulting in differentiation of macrophages that can release cytokines at the second LPS exposure. Therapeutic use of IBET does not re-instate cytokine release in macrophages. (D) Experimental human endotoxemia model, with ex vivo BG administration. Monocytes were isolated from 12 healthy volunteers before (naive) and 4 hr after LPS injection (tolerized). Naive or tolerized monocytes were exposed to BG for 24 hr, followed by culture media, or culture media alone. After 3 days ex vivo, monocytes were re-exposed to LPS, and cytokines were measured 24 hr later. (E and F) BG recovered IL-6 release in 9 out of 12 tolerized monocytes (E) and TNF release in 8 out of 12 monocytes (F). Data are presented as mean ± SD.
Figure 6. Reversal of Tolerance by BG at the Transcriptional Level
(A) Heatmap of the transcriptional response of naive-Mfs, LPS-Mfs, and rescue-Mfs (BG reversed LPS-Mfs) to LPS re-exposure at day 6. The scale represents relative expression between LPS exposed naive-Mfs (1) and LPS-Mfs (0). Rescue-Mfs exposed to LPS show the most similar profile to naive-Mfs. On the top of the heatmap is median expression (log2 RPKM) of tolerized genes at day 6 and LPS re-exposure in naive-Mfs (black), LPS-Mfs (red), LPS-co-IBET-Mfs (blue), LPS + IBET-Mfs (light blue), and rescue-Mfs (purple). (B) BG reverses the tolerization of key LPS-induced transcription factors, such as STAT2, STAT5A, IRF1, and IRF8. Log2 fold change increase in mRNA expression is shown.
Figure 7. Reversal of Tolerance by BG at the Chromatin Level
(A) PCA plot of H3K27ac dynamics among monocytes, naive-Mfs, LPS-Mfs, rescue-Mfs, and LPS-co-IBET-Mfs. BG exposure of tolerized monocytes results in a H3K27ac profile more similar to naive macrophages, while co-incubation of monocytes with LPS and IBET does not lead to activation of these regions. (B) Heatmap showing re-establishment of the naive-Mf H3K27ac signal by BG exposure in tolerized macrophages. (C) PCA plot of H3K4me1 dynamics among monocytes, naive-Mfs, LPS-Mfs, Rescue Mfs, and LPS-co-IBET-Mfs, the effect is similar to H3K27ac, but to a lesser extent. (C) H3K27ac tracks at ATP9B (glucose transport) gene enhancer and (D) LPL (lipid metabolism) gene promoter. See also Figure S7.
Similar articles
- Screening of compounds to identify novel epigenetic regulatory factors that affect innate immune memory in macrophages.
Benjaskulluecha S, Boonmee A, Pattarakankul T, Wongprom B, Klomsing J, Palaga T. Benjaskulluecha S, et al. Sci Rep. 2022 Feb 3;12(1):1912. doi: 10.1038/s41598-022-05929-x. Sci Rep. 2022. PMID: 35115604 Free PMC article. - Immune responsive gene 1 (IRG1) promotes endotoxin tolerance by increasing A20 expression in macrophages through reactive oxygen species.
Li Y, Zhang P, Wang C, Han C, Meng J, Liu X, Xu S, Li N, Wang Q, Shi X, Cao X. Li Y, et al. J Biol Chem. 2013 Jun 7;288(23):16225-16234. doi: 10.1074/jbc.M113.454538. Epub 2013 Apr 22. J Biol Chem. 2013. PMID: 23609450 Free PMC article. - Differential induction of innate memory in porcine monocytes by β-glucan or bacillus Calmette-Guerin.
Byrne KA, Tuggle CK, Loving CL. Byrne KA, et al. Innate Immun. 2021 Aug;27(6):448-460. doi: 10.1177/1753425920951607. Epub 2020 Aug 30. Innate Immun. 2021. PMID: 32862748 Free PMC article. - Molecular mechanisms of innate memory and tolerance to LPS.
Seeley JJ, Ghosh S. Seeley JJ, et al. J Leukoc Biol. 2017 Jan;101(1):107-119. doi: 10.1189/jlb.3MR0316-118RR. Epub 2016 Oct 25. J Leukoc Biol. 2017. PMID: 27780875 Review. - Infectious Agents as Stimuli of Trained Innate Immunity.
Rusek P, Wala M, Druszczyńska M, Fol M. Rusek P, et al. Int J Mol Sci. 2018 Feb 3;19(2):456. doi: 10.3390/ijms19020456. Int J Mol Sci. 2018. PMID: 29401667 Free PMC article. Review.
Cited by
- Myxoma virus lacking the host range determinant M062 stimulates cGAS-dependent type 1 interferon response and unique transcriptomic changes in human monocytes/macrophages.
Conrad SJ, Raza T, Peterson EA, Liem J, Connor R, Nounamo B, Cannon M, Liu J. Conrad SJ, et al. PLoS Pathog. 2022 Sep 14;18(9):e1010316. doi: 10.1371/journal.ppat.1010316. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36103568 Free PMC article. - Innate Immunomodulation in Food Animals: Evidence for Trained Immunity?
Byrne KA, Loving CL, McGill JL. Byrne KA, et al. Front Immunol. 2020 Jun 5;11:1099. doi: 10.3389/fimmu.2020.01099. eCollection 2020. Front Immunol. 2020. PMID: 32582185 Free PMC article. Review. - The Inhibitory Innate Immune Sensor NLRP12 Maintains a Threshold against Obesity by Regulating Gut Microbiota Homeostasis.
Truax AD, Chen L, Tam JW, Cheng N, Guo H, Koblansky AA, Chou WC, Wilson JE, Brickey WJ, Petrucelli A, Liu R, Cooper DE, Koenigsknecht MJ, Young VB, Netea MG, Stienstra R, Sartor RB, Montgomery SA, Coleman RA, Ting JP. Truax AD, et al. Cell Host Microbe. 2018 Sep 12;24(3):364-378.e6. doi: 10.1016/j.chom.2018.08.009. Cell Host Microbe. 2018. PMID: 30212649 Free PMC article. - Epigenetic adjuvants: durable reprogramming of the innate immune system with adjuvants.
Lee A, Wimmers F, Pulendran B. Lee A, et al. Curr Opin Immunol. 2022 Aug;77:102189. doi: 10.1016/j.coi.2022.102189. Epub 2022 May 16. Curr Opin Immunol. 2022. PMID: 35588691 Free PMC article. Review. - Connections between metabolism and epigenetics: mechanisms and novel anti-cancer strategy.
Chen C, Wang Z, Qin Y. Chen C, et al. Front Pharmacol. 2022 Jul 22;13:935536. doi: 10.3389/fphar.2022.935536. eCollection 2022. Front Pharmacol. 2022. PMID: 35935878 Free PMC article. Review.
References
- Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. - PubMed
- Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. - PubMed
- Bahador M, Cross AS. From therapy to experimental model: a hundred years of endotoxin administration to human subjects. J Endotoxin Res. 2007;13:251–279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases