The Future is The Past: Methylation QTLs in Schizophrenia - PubMed (original) (raw)
Review
The Future is The Past: Methylation QTLs in Schizophrenia
Anke Hoffmann et al. Genes (Basel). 2016.
Abstract
Genome-wide association studies (GWAS) have remarkably advanced insight into the genetic basis of schizophrenia (SCZ). Still, most of the functional variance in disease risk remains unexplained. Hence, there is a growing need to map genetic variability-to-genes-to-functions for understanding the pathophysiology of SCZ and the development of better treatments. Genetic variation can regulate various cellular functions including DNA methylation, an epigenetic mark with important roles in transcription and the mediation of environmental influences. Methylation quantitative trait loci (meQTLs) are derived by mapping levels of DNA methylation in genetically different, genotyped individuals and define loci at which DNA methylation is influenced by genetic variation. Recent evidence points to an abundance of meQTLs in brain tissues whose functional contributions to development and mental diseases are still poorly understood. Interestingly, fetal meQTLs reside in regulatory domains affecting methylome reconfiguration during early brain development and are enriched in loci identified by GWAS for SCZ. Moreover, fetal meQTLs are preserved in the adult brain and could trace early epigenomic deregulation during vulnerable periods. Overall, these findings highlight the role of fetal meQTLs in the genetic risk for and in the possible neurodevelopmental origin of SCZ.
Keywords: DNA memory; fetal brain; genome-wide association studies; induced pluripotent stem cells; methylation quantitative trait loci; non-coding variants; schizophrenia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
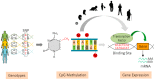
Figure 1
Role of methylation quantitative trait loci (meQTLs) for gene expression. Genetic variation influences levels of DNA methylation at regulatory regions and can modulate gene expression leading to decreased or increased gene transcription. Importantly, the effects of genetically induced changes in DNA methylation critically depend on developmental stage, and possibly, environmental context. SNP: single nucleotide polymorphism.
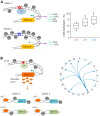
Figure 2
Model for local meQTLs: (A) A local _cis_-acting variant (A-allele symbolized by a red star) resides in a regulatory element, for example a transcription factor (TF) binding site. Sequence variation (G-allele symbolized by a blue star) can result in reduced binding of the TF, diminished transcription of gene A, and encroachment of DNA methylation (left). As depicted by the chart, _cis_-meQTLs show differences in the amount of CpG-methylation (Me) between the two copies of an allele. Homozygous carriers of the transcriptionally active A-allele show less DNA methylation when compared to homozygous carriers of the transcriptionally less active G-allele or heterozygous carriers (right); (B) _Trans_-meQTLs result from differences in the expression, structure, or function of a diffusible factor that is equally available to both alleles at target sites. Accordingly, target sites (genes K and S) do not show differences in allele-specific methylation rates. _Trans_-meQTLs can involve variation in the sequence of TF binding sites (red star) driving expression of the diffusible factor or variation in the coding sequence of the diffusible factor leading to altered structure or function (left). Functionally, _trans_-meQTLs can affect transcription levels of multiple genes. Such trans associations can be shown on a circle plot (chromosomes (chr) labeled 1–22 with arrows pointing to location of a gene on a given chromosome) (right).
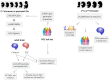
Figure 3
Findings on CpG methylation and on meQTLs in fetal and adult brains from control and SCZ individuals. Results from Jaffe el al. [52] (left) and from Hannon et al. [51] (right) are schematically summarized. In adult brain 62 of 104 Psychiatric Genomics Consortium (PGC) loci harbor a meQTL independent of case control status and fetal meQTLs are four-fold enriched in PGC risk loci. CTCF: CCCTC-binding factor; DBS: DNA binding site.
Similar articles
- Characterization of cross-tissue genetic-epigenetic effects and their patterns in schizophrenia.
Lin D, Chen J, Perrone-Bizzozero N, Bustillo JR, Du Y, Calhoun VD, Liu J. Lin D, et al. Genome Med. 2018 Feb 26;10(1):13. doi: 10.1186/s13073-018-0519-4. Genome Med. 2018. PMID: 29482655 Free PMC article. - Cell-type-specific meQTLs extend melanoma GWAS annotation beyond eQTLs and inform melanocyte gene-regulatory mechanisms.
Zhang T, Choi J, Dilshat R, Einarsdóttir BÓ, Kovacs MA, Xu M, Malasky M, Chowdhury S, Jones K, Bishop DT, Goldstein AM, Iles MM, Landi MT, Law MH, Shi J, Steingrímsson E, Brown KM. Zhang T, et al. Am J Hum Genet. 2021 Sep 2;108(9):1631-1646. doi: 10.1016/j.ajhg.2021.06.018. Epub 2021 Jul 21. Am J Hum Genet. 2021. PMID: 34293285 Free PMC article. - Integrating genome-wide association study and methylation functional annotation data identified candidate genes and pathways for schizophrenia.
Qi X, Guan F, Wen Y, Li P, Ma M, Cheng S, Zhang L, Liang C, Cheng B, Zhang F. Qi X, et al. Prog Neuropsychopharmacol Biol Psychiatry. 2020 Jan 10;96:109736. doi: 10.1016/j.pnpbp.2019.109736. Epub 2019 Aug 16. Prog Neuropsychopharmacol Biol Psychiatry. 2020. PMID: 31425724 - A large-scale integrative analysis of GWAS and common meQTLs across whole life course identifies genes, pathways and tissue/cell types for three major psychiatric disorders.
Zhao Y, Liang X, Zhu F, Wen Y, Xu J, Yang J, Ding M, Cheng B, Ma M, Zhang L, Cheng S, Wu C, Wang S, Wang X, Ning Y, Guo X, Zhang F. Zhao Y, et al. Neurosci Biobehav Rev. 2018 Dec;95:347-352. doi: 10.1016/j.neubiorev.2018.10.005. Epub 2018 Oct 16. Neurosci Biobehav Rev. 2018. PMID: 30339835 Review. - Epigenomics of Major Depressive Disorders and Schizophrenia: Early Life Decides.
Hoffmann A, Sportelli V, Ziller M, Spengler D. Hoffmann A, et al. Int J Mol Sci. 2017 Aug 4;18(8):1711. doi: 10.3390/ijms18081711. Int J Mol Sci. 2017. PMID: 28777307 Free PMC article. Review.
Cited by
- ABCB9 polymorphism rs61955196 is associated with schizophrenia in a Chinese Han population.
Li XW, Zhang MY, Li ZJ, Ai LZ, Jin MD, Jia NN, Xie MT, Yang YQ, Li WZ, Dong L, Yu Q. Li XW, et al. World J Psychiatry. 2022 Jul 19;12(7):904-914. doi: 10.5498/wjp.v12.i7.904. eCollection 2022 Jul 19. World J Psychiatry. 2022. PMID: 36051605 Free PMC article. - Cross-Ancestry DNA Methylation Marks of Insulin Resistance in Pregnancy: An Integrative Epigenome-Wide Association Study.
Fragoso-Bargas N, Elliott HR, Lee-Ødegård S, Opsahl JO, Sletner L, Jenum AK, Drevon CA, Qvigstad E, Moen GH, Birkeland KI, Prasad RB, Sommer C. Fragoso-Bargas N, et al. Diabetes. 2023 Mar 1;72(3):415-426. doi: 10.2337/db22-0504. Diabetes. 2023. PMID: 36534481 Free PMC article. - Moderating effect of mode of delivery on the genetics of intelligence: Explorative genome-wide analyses in ALSPAC.
Smajlagić D, Kvarme Jacobsen K, Myrum C, Haavik J, Johansson S, Zayats T. Smajlagić D, et al. Brain Behav. 2018 Dec;8(12):e01144. doi: 10.1002/brb3.1144. Epub 2018 Oct 31. Brain Behav. 2018. PMID: 30378284 Free PMC article. - Beyond the looking glass: recent advances in understanding the impact of environmental exposures on neuropsychiatric disease.
Hollander JA, Cory-Slechta DA, Jacka FN, Szabo ST, Guilarte TR, Bilbo SD, Mattingly CJ, Moy SS, Haroon E, Hornig M, Levin ED, Pletnikov MV, Zehr JL, McAllister KA, Dzierlenga AL, Garton AE, Lawler CP, Ladd-Acosta C. Hollander JA, et al. Neuropsychopharmacology. 2020 Jun;45(7):1086-1096. doi: 10.1038/s41386-020-0648-5. Epub 2020 Feb 28. Neuropsychopharmacology. 2020. PMID: 32109936 Free PMC article. Review. - Detecting methylation quantitative trait loci using a methylation random field method.
Lyu C, Huang M, Liu N, Chen Z, Lupo PJ, Tycko B, Witte JS, Hobbs CA, Li M. Lyu C, et al. Brief Bioinform. 2021 Nov 5;22(6):bbab323. doi: 10.1093/bib/bbab323. Brief Bioinform. 2021. PMID: 34414410 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous