UniProt: the universal protein knowledgebase - PubMed (original) (raw)
UniProt: the universal protein knowledgebase
The UniProt Consortium. Nucleic Acids Res. 2017.
Erratum in
- UniProt: the universal protein knowledgebase.
UniProt Consortium T. UniProt Consortium T. Nucleic Acids Res. 2018 Mar 16;46(5):2699. doi: 10.1093/nar/gky092. Nucleic Acids Res. 2018. PMID: 29425356 Free PMC article. No abstract available.
Abstract
The UniProt knowledgebase is a large resource of protein sequences and associated detailed annotation. The database contains over 60 million sequences, of which over half a million sequences have been curated by experts who critically review experimental and predicted data for each protein. The remainder are automatically annotated based on rule systems that rely on the expert curated knowledge. Since our last update in 2014, we have more than doubled the number of reference proteomes to 5631, giving a greater coverage of taxonomic diversity. We implemented a pipeline to remove redundant highly similar proteomes that were causing excessive redundancy in UniProt. The initial run of this pipeline reduced the number of sequences in UniProt by 47 million. For our users interested in the accessory proteomes, we have made available sets of pan proteome sequences that cover the diversity of sequences for each species that is found in its strains and sub-strains. To help interpretation of genomic variants, we provide tracks of detailed protein information for the major genome browsers. We provide a SPARQL endpoint that allows complex queries of the more than 22 billion triples of data in UniProt (http://sparql.uniprot.org/). UniProt resources can be accessed via the website at http://www.uniprot.org/.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
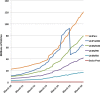
Figure 1.
Growth of the number of sequences in UniProt databases. The blue line shows the growth in UniProtKB/TrEMBL entries from January 2010 to date. The sharp drop in UniProtKB entries corresponds to the proteome redundancy minimization (PRM) procedure implemented in March 2015. Note that the post-PRM growth in UniProtKB is no longer exponential.
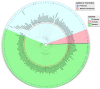
Figure 2.
The distribution of proteomes and reference proteomes across the tree of life.
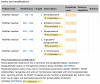
Figure 3.
Screenshot of a part of the ‘PTM/Processing’ section of human TUBA1A entry (UniProtKB Q71U36,
http://www.uniprot.org/uniprot/Q71U36
)
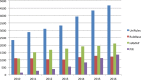
Figure 4.
Growth of automatic annotation rules within UniRule. UniRule integrates rules from HAMAP, PIRSF and RuleBase.
Figure 5.
The GLA gene (P06280, α-galactosidase A) associated with Fabry disease (FD) is shown on the UCSC genome browser using UniProt genome tracks plus variations from ClinVar, dbSNP and OMIM. Panel (A) shows UniProt annotation for a disulfide bond and an amino acid variation associated with FD that removes the Cystene required for a structural fold. Similar situations exist in panel (B) where part of the enzyme's Active Site is disrupted and panel (C) where an N-linked carbohydrate is located. Only the pathogenic variation in C is annotated in other public resources.
Figure 6.
The ProtVista feature viewer. ProtVista uses tracks to display different protein features providing an integrated intuitive picture. The tracks can be expanded, as shown in this Figure with the Variants track. Clicking on a feature highlights its position across all tracks so that co-localized elements can be easily identified. For example here the highlighted site is at the same position as disease correlated natural variants.
Figure 7.
Proteome page for Bacillus subtilis 168. Proteome pages contain a short overview with details about the organism and genome assembly, the list of the genome's components and references from the sequencing projects.
Figure 8.
An example UniRule entry page. Clicking on the conditions highlights the corresponding annotations applied if the conditions hold true and vice versa clicking on the annotations highlights the corresponding conditions. Clicking on the ‘View all proteins annotated by this rule’ button leads to the list of proteins that this rule annotates in UniProtKB.
Figure 9.
The new publications view for UniProtKB entries.
Figure 10.
The Peptide search interface.
Similar articles
- UniProt: the universal protein knowledgebase in 2021.
UniProt Consortium. UniProt Consortium. Nucleic Acids Res. 2021 Jan 8;49(D1):D480-D489. doi: 10.1093/nar/gkaa1100. Nucleic Acids Res. 2021. PMID: 33237286 Free PMC article. - UniProt: the Universal Protein knowledgebase.
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. Apweiler R, et al. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D115-9. doi: 10.1093/nar/gkh131. Nucleic Acids Res. 2004. PMID: 14681372 Free PMC article. - UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View.
Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I. Boutet E, et al. Methods Mol Biol. 2016;1374:23-54. doi: 10.1007/978-1-4939-3167-5_2. Methods Mol Biol. 2016. PMID: 26519399 - In silico characterization of proteins: UniProt, InterPro and Integr8.
Mulder NJ, Kersey P, Pruess M, Apweiler R. Mulder NJ, et al. Mol Biotechnol. 2008 Feb;38(2):165-77. doi: 10.1007/s12033-007-9003-x. Epub 2007 Oct 4. Mol Biotechnol. 2008. PMID: 18219596 Review. - Bioinformatics Tools for Proteomics Data Interpretation.
Calderón-González KG, Hernández-Monge J, Herrera-Aguirre ME, Luna-Arias JP. Calderón-González KG, et al. Adv Exp Med Biol. 2016;919:281-341. doi: 10.1007/978-3-319-41448-5_16. Adv Exp Med Biol. 2016. PMID: 27975225 Review.
Cited by
- Evidence for CAT gene being functionally involved in the susceptibility of COVID-19.
Qian Y, Li Y, Liu X, Yuan N, Ma J, Zheng Q, Liu F. Qian Y, et al. FASEB J. 2021 Apr;35(4):e21384. doi: 10.1096/fj.202100008. FASEB J. 2021. PMID: 33710662 Free PMC article. Review. - Chromosome genome assembly and annotation of Adzuki Bean (Vigna angularis).
Li W, He F, Wang X, Liu Q, Zhang X, Yang Z, Fang C, Xiang H. Li W, et al. Sci Data. 2024 Oct 2;11(1):1074. doi: 10.1038/s41597-024-03911-y. Sci Data. 2024. PMID: 39358398 Free PMC article. - Computational identification of receptor-like kinases "RLK" and receptor-like proteins "RLP" in legumes.
Restrepo-Montoya D, Brueggeman R, McClean PE, Osorno JM. Restrepo-Montoya D, et al. BMC Genomics. 2020 Jul 3;21(1):459. doi: 10.1186/s12864-020-06844-z. BMC Genomics. 2020. PMID: 32620079 Free PMC article. - Analysis of Virus and Host Proteomes During Productive HSV-1 and VZV Infection in Human Epithelial Cells.
Ouwendijk WJD, Dekker LJM, van den Ham HJ, Lenac Rovis T, Haefner ES, Jonjic S, Haas J, Luider TM, Verjans GMGM. Ouwendijk WJD, et al. Front Microbiol. 2020 May 29;11:1179. doi: 10.3389/fmicb.2020.01179. eCollection 2020. Front Microbiol. 2020. PMID: 32547533 Free PMC article. - Sexually Dimorphic Crosstalk at the Maternal-Fetal Interface.
Sun T, Gonzalez TL, Deng N, DiPentino R, Clark EL, Lee B, Tang J, Wang Y, Stripp BR, Yao C, Tseng HR, Karumanchi SA, Koeppel AF, Turner SD, Farber CR, Rich SS, Wang ET, Williams J, Pisarska MD. Sun T, et al. J Clin Endocrinol Metab. 2020 Dec 1;105(12):e4831-47. doi: 10.1210/clinem/dgaa503. J Clin Endocrinol Metab. 2020. PMID: 32772088 Free PMC article.
References
- Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H.. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007; 23:1282–1288. - PubMed
- Leinonen R., Diez F.G., Binns D., Fleischmann W., Lopez R., Apweiler R.. UniProt archive. Bioinformatics. 2004; 20:3236–3237. - PubMed
- Giraldo-Calderon G.I., Emrich S.J., MacCallum R.M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S., VectorBase C., Madey G. et al. . VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–D713. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R13 GM109648/GM/NIGMS NIH HHS/United States
- RG/13/5/30112/BHF_/British Heart Foundation/United Kingdom
- R01 GM080646/GM/NIGMS NIH HHS/United States
- U41 HG002273/HG/NHGRI NIH HHS/United States
- G-1307/PUK_/Parkinson's UK/United Kingdom
- U01 GM120953/GM/NIGMS NIH HHS/United States
- P20 GM103446/GM/NIGMS NIH HHS/United States
- Wellcome Trust/United Kingdom
- U41 HG007822/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources