Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals - PubMed (original) (raw)
Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals
Christian Benedict et al. Mol Metab. 2016.
Abstract
Objective: Changes to the microbial community in the human gut have been proposed to promote metabolic disturbances that also occur after short periods of sleep loss (including insulin resistance). However, whether sleep loss affects the gut microbiota remains unknown.
Methods: In a randomized within-subject crossover study utilizing a standardized in-lab protocol (with fixed meal times and exercise schedules), we studied nine normal-weight men at two occasions: after two nights of partial sleep deprivation (PSD; sleep opportunity 02:45-07:00 h), and after two nights of normal sleep (NS; sleep opportunity 22:30-07:00 h). Fecal samples were collected within 24 h before, and after two in-lab nights, of either NS or PSD. In addition, participants underwent an oral glucose tolerance test following each sleep intervention.
Results: Microbiota composition analysis (V4 16S rRNA gene sequencing) revealed that after two days of PSD vs. after two days of NS, individuals exhibited an increased Firmicutes:Bacteroidetes ratio, higher abundances of the families Coriobacteriaceae and Erysipelotrichaceae, and lower abundance of Tenericutes (all P < 0.05) - previously all associated with metabolic perturbations in animal or human models. However, no PSD vs. NS effect on beta diversity or on fecal short-chain fatty acid concentrations was found. Fasting and postprandial insulin sensitivity decreased after PSD vs. NS (all P < 0.05).
Discussion: Our findings demonstrate that short-term sleep loss induces subtle effects on human microbiota. To what extent the observed changes to the microbial community contribute to metabolic consequences of sleep loss warrants further investigations in larger and more prolonged sleep studies, to also assess how sleep loss impacts the microbiota in individuals who already are metabolically compromised.
Keywords: Bacteroidetes; F:B, Firmicutes:Bacteroidetes (ratio); Firmicutes; HDL, high-density lipoprotein; HOMA-IR, homeostatic assessment model of insulin resistance; Insulin resistance; Intestinal microbiome; LDL, low-density lipoprotein; NS, normal sleep; OGTT, oral glucose tolerance test; OTU, Operational Taxonomic Units; PERMANOVA, permutational analysis of variance; PSD, partial sleep deprivation; SCFA, short-chain fatty acid; Short-chain fatty acid; Sleep restriction; T2DM, type-2 diabetes mellitus; d2, day 2.
Figures
Figure 1
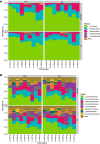
(A) High relative abundance of taxa within the phyla Firmicutes, Actinobacteria, and Bacteroidetes was observed across the fecal samples from the PSD (partial sleep deprivation) and normal sleep (NS) condition. Subjects are arbitrarily numbered and clustered along the y-axis based on bacterial phylum composition. (B) In an analysis across samples at the family level, high abundances were observed of Lachnospiraceae, Ruminococcaceae, Bifidobacteriaceae – and to lesser and more variable extent – of Coriobacteriaceae. (C) The microbiome sequencing analysis of samples obtained after sleep and PSD revealed 136 families; firmicutes was the most diverse phylum, containing the greatest number (69) of the classified families. d2, day 2.
Figure 1
(A) High relative abundance of taxa within the phyla Firmicutes, Actinobacteria, and Bacteroidetes was observed across the fecal samples from the PSD (partial sleep deprivation) and normal sleep (NS) condition. Subjects are arbitrarily numbered and clustered along the y-axis based on bacterial phylum composition. (B) In an analysis across samples at the family level, high abundances were observed of Lachnospiraceae, Ruminococcaceae, Bifidobacteriaceae – and to lesser and more variable extent – of Coriobacteriaceae. (C) The microbiome sequencing analysis of samples obtained after sleep and PSD revealed 136 families; firmicutes was the most diverse phylum, containing the greatest number (69) of the classified families. d2, day 2.
Figure 2
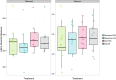
Within and across-subject variation tests for alpha diversity for both conditions (normal sleep, NS; vs. partial sleep deprivation, PSD) and time points (baseline vs. day 2 (d2) sample), using Observed and Shannon methods. See Supplementary Table 2 for statistical comparisons.
Figure 3
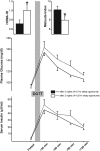
Glucometabolic values in response to normal sleep (black bar and solid lines) and partial sleep deprivation (white bar and dashed lines) for two consecutive nights. HOMA-IR and Matsuda index in the upper panel were obtained in the fasting state and from an oral glucose tolerance test (OGTT), respectively. Curves for plasma glucose (middle panel) and insulin (lower panel) were obtained from pre and post (up to 120 min) OGTT values. n = 9; *, P < 0.05.
Comment in
- Sleepy, circadian disrupted and sick: Could intestinal microbiota play an important role in shift worker health?
Reynolds AC, Broussard J, Paterson JL, Wright KP Jr, Ferguson SA. Reynolds AC, et al. Mol Metab. 2016 Nov 21;6(1):12-13. doi: 10.1016/j.molmet.2016.11.004. eCollection 2017 Jan. Mol Metab. 2016. PMID: 28123932 Free PMC article. No abstract available.
Similar articles
- The stool microbiota of insulin resistant women with recent gestational diabetes, a high risk group for type 2 diabetes.
Fugmann M, Breier M, Rottenkolber M, Banning F, Ferrari U, Sacco V, Grallert H, Parhofer KG, Seissler J, Clavel T, Lechner A. Fugmann M, et al. Sci Rep. 2015 Aug 17;5:13212. doi: 10.1038/srep13212. Sci Rep. 2015. PMID: 26279179 Free PMC article. - [Analysis of the dynamic changes in gut microbiota in patients with extremely severe burns by 16S ribosomal RNA high-throughput sequencing technology].
Pan YY, Fan YF, Li JL, Cui SY, Huang N, Jin GY, Chen C, Zhang C. Pan YY, et al. Zhonghua Shao Shang Za Zhi. 2020 Dec 20;36(12):1159-1166. doi: 10.3760/cma.j.cn501120-20200518-00271. Zhonghua Shao Shang Za Zhi. 2020. PMID: 33379852 Chinese. - Characterization of the Gut Microbial Community of Obese Patients Following a Weight-Loss Intervention Using Whole Metagenome Shotgun Sequencing.
Louis S, Tappu RM, Damms-Machado A, Huson DH, Bischoff SC. Louis S, et al. PLoS One. 2016 Feb 26;11(2):e0149564. doi: 10.1371/journal.pone.0149564. eCollection 2016. PLoS One. 2016. PMID: 26919743 Free PMC article. - Systematic review assessing the effectiveness of dietary intervention on gut microbiota in adults with type 2 diabetes.
Houghton D, Hardy T, Stewart C, Errington L, Day CP, Trenell MI, Avery L. Houghton D, et al. Diabetologia. 2018 Aug;61(8):1700-1711. doi: 10.1007/s00125-018-4632-0. Epub 2018 May 12. Diabetologia. 2018. PMID: 29754286 Free PMC article. - The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients?
Magne F, Gotteland M, Gauthier L, Zazueta A, Pesoa S, Navarrete P, Balamurugan R. Magne F, et al. Nutrients. 2020 May 19;12(5):1474. doi: 10.3390/nu12051474. Nutrients. 2020. PMID: 32438689 Free PMC article. Review.
Cited by
- Is Sleep Associated with the S-Klotho Anti-Aging Protein in Sedentary Middle-Aged Adults? The FIT-AGEING Study.
Mochón-Benguigui S, Carneiro-Barrera A, Castillo MJ, Amaro-Gahete FJ. Mochón-Benguigui S, et al. Antioxidants (Basel). 2020 Aug 12;9(8):738. doi: 10.3390/antiox9080738. Antioxidants (Basel). 2020. PMID: 32806634 Free PMC article. - Caffeine-Induced Sleep Restriction Alters the Gut Microbiome and Fecal Metabolic Profiles in Mice.
Song Z, Liu L, Xu Y, Cao R, Lan X, Pan C, Zhang S, Zhao H. Song Z, et al. Int J Mol Sci. 2022 Nov 27;23(23):14837. doi: 10.3390/ijms232314837. Int J Mol Sci. 2022. PMID: 36499163 Free PMC article. - A brief period of sleep deprivation leads to subtle changes in mouse gut microbiota.
El Aidy S, Bolsius YG, Raven F, Havekes R. El Aidy S, et al. J Sleep Res. 2020 Dec;29(6):e12920. doi: 10.1111/jsr.12920. Epub 2019 Sep 12. J Sleep Res. 2020. PMID: 31515894 Free PMC article. - Current understanding of the human microbiome.
Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Gilbert JA, et al. Nat Med. 2018 Apr 10;24(4):392-400. doi: 10.1038/nm.4517. Nat Med. 2018. PMID: 29634682 Free PMC article. Review. - Sleep and the gut microbiota in preschool-aged children.
Wang Y, van de Wouw M, Drogos L, Vaghef-Mehrabani E, Reimer RA, Tomfohr-Madsen L, Giesbrecht GF. Wang Y, et al. Sleep. 2022 Jun 13;45(6):zsac020. doi: 10.1093/sleep/zsac020. Sleep. 2022. PMID: 35037059 Free PMC article.
References
- Vrieze A., Van Nood E., Holleman F., Salojarvi J., Kootte R.S., Bartelsman J.F. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–916.e7. - PubMed
- Hartstra A.V., Bouter K.E., Backhed F., Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. 2015;38(1):159–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous