Single-cell studies of IFN-β promoter activation by wild-type and NS1-defective influenza A viruses - PubMed (original) (raw)
Single-cell studies of IFN-β promoter activation by wild-type and NS1-defective influenza A viruses
M J Killip et al. J Gen Virol. 2017 Mar.
Abstract
Deletion or truncation of NS1, the principal IFN antagonist of influenza viruses, leads to increased IFN induction during influenza virus infection. We have studied activation of the IFN induction cascade by both wild-type and NS1-defective viruses at the single-cell level using a cell line expressing GFP under the control of the IFN-β promoter and by examining MxA expression. The IFN-β promoter was not activated in all infected cells even during NS1-defective virus infections. Loss of NS1 expression is therefore insufficient per se to induce IFN in an infected cell, and factors besides NS1 expression status must dictate whether the IFN response is activated. The IFN response was efficiently stimulated in these cells following infection with other viruses; the differential IFN response we observe with influenza viruses is therefore not cell specific but is likely due to differences in the nature of the infecting virus particles and their subsequent replication.
Figures
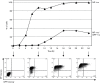
Fig. 1.
Failure to activate the IFN-β promoter by NS1-defective influenza A viruses. (a) A549/pr(IFN-β).GFP cells were uninfected or infected with Ud wt, PR8 wt or Sendai virus (SeV) Cantell at 5 p.f.u. cell−1. At 16 h post-infection (p.i.), cells were trypsinized, fixed, permeabilized and immunostained with antibody against influenza virus NP and subsequently analysed for NP and GFP expression by flow cytometry. Cells were divided into quadrants according to intensity of NP and GFP expression, and the percentage of cells in each quadrant is indicated on each graph. (b) Cell lysates were generated from A549/pr(IFN-β).GFP monolayers infected with Ud wt, Ud-Δ99R38A, PR8 wt or PR8-ΔNS1 at 5 p.f.u. cell−1 or uninfected cells for 16 h p.i., then subjected to SDS-PAGE and immunoblotting with antibodies specific to phospho-IRF3, GFP, viral proteins and actin. (c) Cells were treated as in (b). IFN present in culture media was estimated by a cytopathic effect-reduction bioassay [31]. Error bars represent the results of three independent experiments. (d, e) A549/pr(IFN-β).GFP cells were uninfected or infected with Ud-Δ99/R38A or PR8-ΔNS1 at the multiplicities indicated on the plots. At 16 h p.i., cells were trypsinized, fixed, permeabilized and immunostained for influenza virus haemagglutinin or NP expression as indicated. PIV5-VΔC vM2 [32] or SeV Cantell infections were also carried out as positive controls for GFP expression. Cells were analysed by flow cytometry as in (a). (f) Cells were infected as in (d) and (e). At 16 h p.i., cells were fixed, permeabilized and immunostained for influenza virus NP. GFP, NP (red) and nuclei (stained with DAPI; grey) were visualized by confocal microscopy. Arrows denote those cells that are strongly positive for virus antigen but in which GFP cannot be detected.
Fig. 1.
Failure to activate the IFN-β promoter by NS1-defective influenza A viruses. (a) A549/pr(IFN-β).GFP cells were uninfected or infected with Ud wt, PR8 wt or Sendai virus (SeV) Cantell at 5 p.f.u. cell−1. At 16 h post-infection (p.i.), cells were trypsinized, fixed, permeabilized and immunostained with antibody against influenza virus NP and subsequently analysed for NP and GFP expression by flow cytometry. Cells were divided into quadrants according to intensity of NP and GFP expression, and the percentage of cells in each quadrant is indicated on each graph. (b) Cell lysates were generated from A549/pr(IFN-β).GFP monolayers infected with Ud wt, Ud-Δ99R38A, PR8 wt or PR8-ΔNS1 at 5 p.f.u. cell−1 or uninfected cells for 16 h p.i., then subjected to SDS-PAGE and immunoblotting with antibodies specific to phospho-IRF3, GFP, viral proteins and actin. (c) Cells were treated as in (b). IFN present in culture media was estimated by a cytopathic effect-reduction bioassay [31]. Error bars represent the results of three independent experiments. (d, e) A549/pr(IFN-β).GFP cells were uninfected or infected with Ud-Δ99/R38A or PR8-ΔNS1 at the multiplicities indicated on the plots. At 16 h p.i., cells were trypsinized, fixed, permeabilized and immunostained for influenza virus haemagglutinin or NP expression as indicated. PIV5-VΔC vM2 [32] or SeV Cantell infections were also carried out as positive controls for GFP expression. Cells were analysed by flow cytometry as in (a). (f) Cells were infected as in (d) and (e). At 16 h p.i., cells were fixed, permeabilized and immunostained for influenza virus NP. GFP, NP (red) and nuclei (stained with DAPI; grey) were visualized by confocal microscopy. Arrows denote those cells that are strongly positive for virus antigen but in which GFP cannot be detected.
Fig. 2.
Timecourse of GFP expression in A549/pr(IFN-β).GFP cells during infection with an NS1-defective influen
z
a A virus. A549/pr(IFN-β).GFP cells were infected with Ud-Δ99/R38A at 5 p.f.u. cell−1. At the indicated times post-infection, cells were trypsinized, fixed, permeabilized and immunostained for NP expression. GFP and NP expression were subsequently analysed by flow cytometry. The percentage of cells positive for NP and the percentage of cells positive for both NP and GFP at each timepoint are plotted. Flow cytometry plots at selected timepoints are shown below the graph.
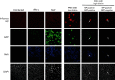
Fig. 3.
Heterogeneity in the induction of an antiviral state in uninfected cells surrounding NS1-defective influenza A virus-infected cells. A549/pr(IFN-β).GFP cells were infected with low or high dilutions of PR8-ΔNS1 as indicated, uninfected or infected with SeV Cantell as a positive control for GFP expression. As a positive control for MxA expression, cells were treated with IFN-α (1000 IU ml−1). Cells were fixed at 16 h p.i., permeabilized and immunostained for influenza virus NP and MxA. Nuclear material was stained with DAPI. GFP, MxA, NP and DAPI staining were examined by confocal microscopy. Arrows denote the positions of NP-positive cells that are either GFP positive and surrounded by MxA-positive cells or are GFP negative and surrounded by MxA-negative cells.
Similar articles
- Generation of replication-proficient influenza virus NS1 point mutants with interferon-hyperinducer phenotype.
Pérez-Cidoncha M, Killip MJ, Asensio VJ, Fernández Y, Bengoechea JA, Randall RE, Ortín J. Pérez-Cidoncha M, et al. PLoS One. 2014 Jun 2;9(6):e98668. doi: 10.1371/journal.pone.0098668. eCollection 2014. PLoS One. 2014. PMID: 24887174 Free PMC article. - Regulation of interferon-β by MAGI-1 and its interaction with influenza A virus NS1 protein with ESEV PBM.
Kumar M, Liu H, Rice AP. Kumar M, et al. PLoS One. 2012;7(7):e41251. doi: 10.1371/journal.pone.0041251. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911767 Free PMC article. - Quantitative proteomic analysis of the influenza A virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor.
Tawaratsumida K, Phan V, Hrincius ER, High AA, Webby R, Redecke V, Häcker H. Tawaratsumida K, et al. J Virol. 2014 Aug;88(16):9038-48. doi: 10.1128/JVI.00830-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899174 Free PMC article. - Functions of the influenza A virus NS1 protein in antiviral defense.
Krug RM. Krug RM. Curr Opin Virol. 2015 Jun;12:1-6. doi: 10.1016/j.coviro.2015.01.007. Epub 2015 Jan 29. Curr Opin Virol. 2015. PMID: 25638592 Free PMC article. Review. - Research progress on the nonstructural protein 1 (NS1) of influenza a virus.
Zhang X, Zhang Y, Wei F. Zhang X, et al. Virulence. 2024 Dec;15(1):2359470. doi: 10.1080/21505594.2024.2359470. Epub 2024 Jun 25. Virulence. 2024. PMID: 38918890 Free PMC article. Review.
Cited by
- Maximal interferon induction by influenza lacking NS1 is infrequent owing to requirements for replication and export.
Vicary AC, Mendes M, Swaminath S, Lekbua A, Reddan J, Rodriguez ZK, Russell AB. Vicary AC, et al. PLoS Pathog. 2023 Apr 17;19(4):e1010943. doi: 10.1371/journal.ppat.1010943. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37068114 Free PMC article. - Defective Interfering Particles of Influenza Virus and Their Characteristics, Impacts, and Use in Vaccines and Antiviral Strategies: A Systematic Review.
Wu M, Zhou E, Sheng R, Fu X, Li J, Jiang C, Su W. Wu M, et al. Viruses. 2022 Dec 12;14(12):2773. doi: 10.3390/v14122773. Viruses. 2022. PMID: 36560777 Free PMC article. Review. - Comprehensive single cell analysis of pandemic influenza A virus infection in the human airways uncovers cell-type specific host transcriptional signatures relevant for disease progression and pathogenesis.
Kelly JN, Laloli L, V'kovski P, Holwerda M, Portmann J, Thiel V, Dijkman R. Kelly JN, et al. Front Immunol. 2022 Oct 4;13:978824. doi: 10.3389/fimmu.2022.978824. eCollection 2022. Front Immunol. 2022. PMID: 36268025 Free PMC article. - Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of IFN-mediated innate immune defenses.
Herder V, Dee K, Wojtus JK, Epifano I, Goldfarb D, Rozario C, Gu Q, Da Silva Filipe A, Nomikou K, Nichols J, Jarrett RF, Stevenson A, McFarlane S, Stewart ME, Szemiel AM, Pinto RM, Masdefiol Garriga A, Davis C, Allan J, Graham SV, Murcia PR, Boutell C. Herder V, et al. PLoS Biol. 2021 Dec 21;19(12):e3001065. doi: 10.1371/journal.pbio.3001065. eCollection 2021 Dec. PLoS Biol. 2021. PMID: 34932557 Free PMC article. - Library-based analysis reveals segment and length dependent characteristics of defective influenza genomes.
Mendes M, Russell AB. Mendes M, et al. PLoS Pathog. 2021 Dec 9;17(12):e1010125. doi: 10.1371/journal.ppat.1010125. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34882752 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical