The cornucopia of meaningful leads: Applying deep adversarial autoencoders for new molecule development in oncology - PubMed (original) (raw)
The cornucopia of meaningful leads: Applying deep adversarial autoencoders for new molecule development in oncology
Artur Kadurin et al. Oncotarget. 2017.
Abstract
Recent advances in deep learning and specifically in generative adversarial networks have demonstrated surprising results in generating new images and videos upon request even using natural language as input. In this paper we present the first application of generative adversarial autoencoders (AAE) for generating novel molecular fingerprints with a defined set of parameters. We developed a 7-layer AAE architecture with the latent middle layer serving as a discriminator. As an input and output the AAE uses a vector of binary fingerprints and concentration of the molecule. In the latent layer we also introduced a neuron responsible for growth inhibition percentage, which when negative indicates the reduction in the number of tumor cells after the treatment. To train the AAE we used the NCI-60 cell line assay data for 6252 compounds profiled on MCF-7 cell line. The output of the AAE was used to screen 72 million compounds in PubChem and select candidate molecules with potential anti-cancer properties. This approach is a proof of concept of an artificially-intelligent drug discovery engine, where AAEs are used to generate new molecular fingerprints with the desired molecular properties.
Keywords: adversarial autoencoder; artificial intelligence; deep learning; drug discovery; generative adversarian networks.
Conflict of interest statement
CONFLICTS OF INTEREST
The authors are affiliated with Insilico Medicine, Inc, which is applying deep learning techniques, including generative adversarial networks to drug discovery for internal research, providing services to the pharmaceutical companies and identifying novel geroprotectors. The authors have vested interest in demonstrating and popularizing the successful applications of the deep learning techniques for drug discovery and biomarker development.
Figures
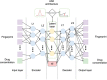
Figure 1. Architecture of Adversarial Autoencoder (AAE) used in this study
Encoder consists of two consequent layers L1 and L2 with 128 and 64 neurons, respectively. In turn, decoder consists of layers L'1 and L'2 comprising 64 and 128 neurons. Latent layer consists of 5 neurons one of which is Growth Inhibition percentage (GI) and the other 4 are discriminated with normal distribution.
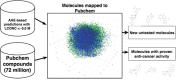
Figure 2. Mapping generated molecules to chemical space of Pubchem
Pubchem compounds are depicted in green, training set is shown in blue and mapped predictions in red.
Similar articles
- druGAN: An Advanced Generative Adversarial Autoencoder Model for de Novo Generation of New Molecules with Desired Molecular Properties in Silico.
Kadurin A, Nikolenko S, Khrabrov K, Aliper A, Zhavoronkov A. Kadurin A, et al. Mol Pharm. 2017 Sep 5;14(9):3098-3104. doi: 10.1021/acs.molpharmaceut.7b00346. Epub 2017 Aug 4. Mol Pharm. 2017. PMID: 28703000 - Deep Convolutional Generative Adversarial Network (dcGAN) Models for Screening and Design of Small Molecules Targeting Cannabinoid Receptors.
Bian Y, Wang J, Jun JJ, Xie XQ. Bian Y, et al. Mol Pharm. 2019 Nov 4;16(11):4451-4460. doi: 10.1021/acs.molpharmaceut.9b00500. Epub 2019 Oct 24. Mol Pharm. 2019. PMID: 31589460 - Adversarial deep evolutionary learning for drug design.
Abouchekeir S, Vu A, Mukaidaisi M, Grantham K, Tchagang A, Li Y. Abouchekeir S, et al. Biosystems. 2022 Dec;222:104790. doi: 10.1016/j.biosystems.2022.104790. Epub 2022 Oct 11. Biosystems. 2022. PMID: 36228831 - Generative machine learning for de novo drug discovery: A systematic review.
Martinelli DD. Martinelli DD. Comput Biol Med. 2022 Jun;145:105403. doi: 10.1016/j.compbiomed.2022.105403. Epub 2022 Mar 13. Comput Biol Med. 2022. PMID: 35339849 Review. - The power of deep learning to ligand-based novel drug discovery.
Baskin II. Baskin II. Expert Opin Drug Discov. 2020 Jul;15(7):755-764. doi: 10.1080/17460441.2020.1745183. Epub 2020 Mar 31. Expert Opin Drug Discov. 2020. PMID: 32228116 Review.
Cited by
- Predicting Aging of Brain Metabolic Topography Using Variational Autoencoder.
Choi H, Kang H, Lee DS; Alzheimer's Disease Neuroimaging Initiative. Choi H, et al. Front Aging Neurosci. 2018 Jul 12;10:212. doi: 10.3389/fnagi.2018.00212. eCollection 2018. Front Aging Neurosci. 2018. PMID: 30050430 Free PMC article. - cheML.io: an online database of ML-generated molecules.
Zhumagambetov R, Kazbek D, Shakipov M, Maksut D, Peshkov VA, Fazli S. Zhumagambetov R, et al. RSC Adv. 2020 Dec 22;10(73):45189-45198. doi: 10.1039/d0ra07820d. eCollection 2020 Dec 17. RSC Adv. 2020. PMID: 35516285 Free PMC article. - Machine and deep learning approaches for cancer drug repurposing.
Issa NT, Stathias V, Schürer S, Dakshanamurthy S. Issa NT, et al. Semin Cancer Biol. 2021 Jan;68:132-142. doi: 10.1016/j.semcancer.2019.12.011. Epub 2020 Jan 3. Semin Cancer Biol. 2021. PMID: 31904426 Free PMC article. Review. - Accelerating Therapeutics for Opportunities in Medicine: A Paradigm Shift in Drug Discovery.
Hinkson IV, Madej B, Stahlberg EA. Hinkson IV, et al. Front Pharmacol. 2020 Jun 30;11:770. doi: 10.3389/fphar.2020.00770. eCollection 2020. Front Pharmacol. 2020. PMID: 32694991 Free PMC article.
References
- Munos BH, Chin WW. How to revive breakthrough innovation in the pharmaceutical industry. Sci Transl Med. 2011;3:89cm16. - PubMed
- Mignani S, Huber S, Tomás H, Rodrigues J, Majoral J-P. Why and how have drug discovery strategies in pharma changed? What are the new mindsets? Drug Discov Today. 2016;21:239–49. - PubMed
- Kola I, Ismail K, John L. Opinion: Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–6. - PubMed
- Thomas DW, Burns J, Audette J, Carrol A, Dow-Hygelund C, Hay M. San Diego: Biomedtracker/Washington, DC: BIO/Bend: Amplion; 2016. Clinical development success rates 2006-2015.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials