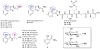Identification of Neuroprotective Spoxazomicin and Oxachelin Glycosides via Chemoenzymatic Glycosyl-Scanning - PubMed (original) (raw)
. 2017 Jan 27;80(1):12-18.
doi: 10.1021/acs.jnatprod.6b00949. Epub 2016 Dec 28.
Ryan R Hughes 1, Meredith A Saunders 1, Sherif I Elshahawi 1, Larissa V Ponomareva 1, Yinan Zhang 1, Sydney R Winchester 1, Samantha A Scott 1, Manjula Sunkara 1, Andrew J Morris 1, Mark A Prendergast 1, Khaled A Shaaban 1, Jon S Thorson 1
Affiliations
- PMID: 28029796
- PMCID: PMC5337260
- DOI: 10.1021/acs.jnatprod.6b00949
Identification of Neuroprotective Spoxazomicin and Oxachelin Glycosides via Chemoenzymatic Glycosyl-Scanning
Jianjun Zhang et al. J Nat Prod. 2017.
Abstract
The assessment of glycosyl-scanning to expand the molecular and functional diversity of metabolites from the underground coal mine fire-associated Streptomyces sp. RM-14-6 is reported. Using the engineered glycosyltransferase OleD Loki and a 2-chloro-4-nitrophenylglycoside-based screen, six metabolites were identified as substrates of OleD Loki, from which 12 corresponding metabolite glycosides were produced and characterized. This study highlights the first application of the 2-chloro-4-nitrophenylglycoside-based screen toward an unbiased set of unique microbial natural products and the first reported application of the 2-chloro-4-nitrophenylglycoside-based transglycosylation reaction for the corresponding preparative synthesis of target glycosides. Bioactivity analysis (including antibacterial, antifungal, anticancer, and EtOH damage neuroprotection assays) revealed glycosylation to attenuate the neuroprotective potency of 4, while glycosylation of the structurally related inactive spoxazomicin C (3) remarkably invoked neuroprotective activity.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors report competing interests. JST is a co-founder of Centrose (Madison, WI).
Figures
Figure 1
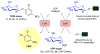
General glycosyl-scanning scheme. In this strategy, activated 2-chloro-4-nitrophenylglycoside donors help drive the OleD Loki-catalyzed transglycosylation reaction toward desired product formation where the resulting production of colorimetric 2-chloro-4-nitrophenol affords a real-time indicator of sugar-nucleotide production and utilization as an indirect measure of target glycoside production.
Figure 2
Structures of validated OleD Loki substrates identified and corresponding glycosides generated via preparative OleD Loki-catalyzed chemoenzymatic synthesis.
Figure 3
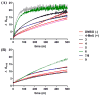
OleD Loki-catalyzed colorimetric glycosyl-scanning results using 2-chloro-4-nitrophenyl-Glc (1) (A) or 2-chloro-4-nitrophenyl-GlcNH2 (2) (B) as donor for all Streptomyces sp. RM-14-6 metabolites confirmed to turnover based on LC-MS [positive control, 4-methylumbelliferone (4-MeU); negative control, no acceptor (DMSO)]. No turnover was observed with compounds 10–20 (Supporting Information, Figure S1). The turnover of compound 8 (the synthetic enantiomer of metabolite 7) was identical to 7.
Figure 4
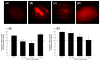
EtOH damage neuroprotection assay (propidium iodide uptake in rat-derived organotypic hippocampal slice primary cell cultures). (A) DMSO control; (B) 48 h exposure to 100 mM EtOH; (C) 48 h exposure to 100 mM EtOH and 10 nM 3a; (D) 48 h exposure to 100 mM EtOH and 1 _μ_M 4a; (E) dose–response with 48 h exposure to EtOH (100 mM) in the absence or presence of 3a on propidium iodide uptake; (F) Ddose–response with 48 h exposure to EtOH (100 mM) in the absence or presence of 4a on propidium iodide uptake. *p < 0.001 vs control; **p < 0.001 vs EtOH.
Similar articles
- Spoxazomicin D and Oxachelin C, Potent Neuroprotective Carboxamides from the Appalachian Coal Fire-Associated Isolate Streptomyces sp. RM-14-6.
Shaaban KA, Saunders MA, Zhang Y, Tran T, Elshahawi SI, Ponomareva LV, Wang X, Zhang J, Copley GC, Sunkara M, Kharel MK, Morris AJ, Hower JC, Tremblay MS, Prendergast MA, Thorson JS. Shaaban KA, et al. J Nat Prod. 2017 Jan 27;80(1):2-11. doi: 10.1021/acs.jnatprod.6b00948. Epub 2016 Dec 28. J Nat Prod. 2017. PMID: 28029795 Free PMC article. - Oxachelin, a novel iron chelator and antifungal agent from Streptomyces sp. GW9/1258.
Sontag B, Gerlitz M, Paululat T, Rasser HF, Grün-Wollny I, Hansske FG. Sontag B, et al. J Antibiot (Tokyo). 2006 Oct;59(10):659-63. doi: 10.1038/ja.2006.88. J Antibiot (Tokyo). 2006. PMID: 17191682 - Terfestatins B and C, New p-Terphenyl Glycosides Produced by Streptomyces sp. RM-5-8.
Wang X, Reynolds AR, Elshahawi SI, Shaaban KA, Ponomareva LV, Saunders MA, Elgumati IS, Zhang Y, Copley GC, Hower JC, Sunkara M, Morris AJ, Kharel MK, Van Lanen SG, Prendergast MA, Thorson JS. Wang X, et al. Org Lett. 2015 Jun 5;17(11):2796-9. doi: 10.1021/acs.orglett.5b01203. Epub 2015 May 11. Org Lett. 2015. PMID: 25961722 Free PMC article. - Flavonoids as Aglycones in Retaining Glycosidase-Catalyzed Reactions: Prospects for Green Chemistry.
Kotik M, Kulik N, Valentová K. Kotik M, et al. J Agric Food Chem. 2023 Oct 18;71(41):14890-14910. doi: 10.1021/acs.jafc.3c04389. Epub 2023 Oct 6. J Agric Food Chem. 2023. PMID: 37800688 Free PMC article. Review. - Design of chemical glycosyl donors: does changing ring conformation influence selectivity/reactivity?
Satoh H, Manabe S. Satoh H, et al. Chem Soc Rev. 2013 May 21;42(10):4297-309. doi: 10.1039/c3cs35457a. Epub 2013 Jan 30. Chem Soc Rev. 2013. PMID: 23364773 Review.
Cited by
- Structural Reassignment and Absolute Stereochemistry of Madurastatin C1 (MBJ-0034) and the Related Aziridine Siderophores: Madurastatins A1, B1, and MBJ-0035.
Tyler AR, Mosaei H, Morton S, Waddell PG, Wills C, McFarlane W, Gray J, Goodfellow M, Errington J, Allenby N, Zenkin N, Hall MJ. Tyler AR, et al. J Nat Prod. 2017 May 26;80(5):1558-1562. doi: 10.1021/acs.jnatprod.7b00082. Epub 2017 Apr 11. J Nat Prod. 2017. PMID: 28398740 Free PMC article. - Sugar-Pirating as an Enabling Platform for the Synthesis of 4,6-Dideoxyhexoses.
Zhang Y, Zhang J, Ponomareva LV, Cui Z, Van Lanen SG, Thorson JS. Zhang Y, et al. J Am Chem Soc. 2020 May 20;142(20):9389-9395. doi: 10.1021/jacs.9b13766. Epub 2020 May 7. J Am Chem Soc. 2020. PMID: 32330028 Free PMC article. - Modulating the Serotonin Receptor Spectrum of Pulicatin Natural Products.
Lin Z, Smith MD, Concepcion GP, Haygood MG, Olivera BM, Light A, Schmidt EW. Lin Z, et al. J Nat Prod. 2017 Aug 25;80(8):2360-2370. doi: 10.1021/acs.jnatprod.7b00317. Epub 2017 Jul 26. J Nat Prod. 2017. PMID: 28745513 Free PMC article. - OleD Loki as a Catalyst for Hydroxamate Glycosylation.
Hughes RR, Shaaban KA, Ponomareva LV, Horn J, Zhang C, Zhan CG, Voss SR, Leggas M, Thorson JS. Hughes RR, et al. Chembiochem. 2020 Apr 1;21(7):952-957. doi: 10.1002/cbic.201900601. Epub 2019 Nov 26. Chembiochem. 2020. PMID: 31621997 Free PMC article.
References
- Moloney MG. Trends Pharmacol Sci. 2016;37:689–701. - PubMed
- Harvey AL, Edrada-Ebel R, Quinn RJ. Nat Rev Drug Discovery. 2015;14:111–129. - PubMed
- Cragg GM, Grothaus PG, Newman DJ. J Nat Prod. 2014;77:703–723. - PubMed
- Li JW, Vederas JC. Science. 2009;325:161–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 TR000117/TR/NCATS NIH HHS/United States
- R37 AI052218/AI/NIAID NIH HHS/United States
- T32 DA016176/DA/NIDA NIH HHS/United States
- UL1 TR001998/TR/NCATS NIH HHS/United States
- P20 GM103527/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources