Progress in Understanding the Molecular Basis Underlying Functional Diversification of Cyclic Dinucleotide Turnover Proteins - PubMed (original) (raw)
Review
. 2017 Feb 14;199(5):e00790-16.
doi: 10.1128/JB.00790-16. Print 2017 Mar 1.
Affiliations
- PMID: 28031279
- PMCID: PMC5309910
- DOI: 10.1128/JB.00790-16
Review
Progress in Understanding the Molecular Basis Underlying Functional Diversification of Cyclic Dinucleotide Turnover Proteins
Ute Römling et al. J Bacteriol. 2017.
Abstract
Cyclic di-GMP was the first cyclic dinucleotide second messenger described, presaging the discovery of additional cyclic dinucleotide messengers in bacteria and eukaryotes. The GGDEF diguanylate cyclase (DGC) and EAL and HD-GYP phosphodiesterase (PDE) domains conduct the turnover of cyclic di-GMP. These three unrelated domains belong to superfamilies that exhibit significant variations in function, and they include both enzymatically active and inactive members, with a subset involved in synthesis and degradation of other cyclic dinucleotides. Here, we summarize current knowledge of sequence and structural variations that underpin the functional diversification of cyclic di-GMP turnover proteins. Moreover, we highlight that superfamily diversification is not restricted to cyclic di-GMP signaling domains, as particular DHH/DHHA1 domain and HD domain proteins have been shown to act as cyclic di-AMP phosphodiesterases. We conclude with a consideration of the current limitations that such diversity of action places on bioinformatic prediction of the roles of GGDEF, EAL, and HD-GYP domain proteins.
Keywords: DHH/DHHA1 domain; EAL domain; GGDEF domain; HD-GYP domain; cyclic GAMP; cyclic di-AMP; cyclic di-GMP; cyclic dinucleotide second messenger; second messenger.
Copyright © 2017 American Society for Microbiology.
Figures
FIG 1
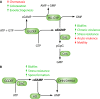
Enzymes involved in the turnover of second messengers cyclic di-GMP (A) and cyclic di-AMP (B). GGDEF domain proteins are cyclic di-GMP synthases (2), but a few members can preferentially synthesize cyclic GMP-AMP (31). Cyclic di-GMP is degraded by EAL domain and HD-GYP domain phosphodiesterases into 5′-pGpG and GMP, respectively (66). 5′-pGpG is further hydrolyzed to GMP by the oligoribonuclease Orn. Cyclic di-AMP is synthesized by the DAC domain, and hydrolysis by HD and DHH/DHHA1 domain proteins has been demonstrated. Major phenotypes affected upon cyclic dinucleotide synthesis in many bacteria are indicated.
FIG 2
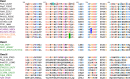
Classification of GGDEF domains according to protein structure and conservation of signature motifs. Amino acids on a gray background interact with the substrate in the diguanylate cyclase PleD. K332, stabilizing the transition state, is on a cyan background. The RXXD I-site core motif is in blue. Unconventional amino acids still conferring enzymatic activity are on a blue background. Amino acids conferring cyclic GMP-AMP specificity are on a green background. Amino acids involved in protein-protein interactions are underlined. Conserved amino acids are color coded. GGDEF domain protein names are in black, and GGDEF-EAL proteins are in green. Unconventional GGDEF domain names are in violet, and cGMP-AMP-synthesizing proteins are in orange. Protein designations are given in the supplemental material. Modified from the work of Römling et al. (32).
FIG 3
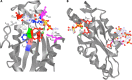
Ribbon diagram of the GGDEF domain of PleD binding the substrate analog GTPαS (PDB code
2V0N
). (A) Amino acids interacting with the substrate analog GTPαS (including Lys442 and Arg446, interacting with the phosphate group) and the Mg2+ ions (Asp327 and Glu370) (1, 6, 15) are indicated. Mg2+ ions are in green. (B) Amino acid motifs providing additional functionality to GGDEF domains. R359XXD362 is the core motif of the I-site. The XXD327XD329 motif was demonstrated to have a role in protein-protein interactions in GGDEF domain proteins others than PleD.
FIG 4
Classification of EAL domains according to protein structure and conservation of signature motifs. The catalytic base glutamate is shown in red. Green, amino acids involved in Mg2+ binding; blue, amino acids involved in substrate binding. The glutamate-stabilizing loop 6 is shown in orange. Loop 6 amino acids are on a gray background. Mutated loop 6 amino acids in class I PleD, loop 6 amino acids determinative for lack of catalytic activity of the EAL domain in DGC2_Komyxl, and alternative amino acids involved in cyclic dinucleotide binding in class IIIA proteins are underlined. Names of EAL proteins are in black, EAL-only proteins are in red, and GGDEF-EAL proteins are in green. Protein designations are given in the supplemental material. Modified from the work of Römling et al. (32).
FIG 5
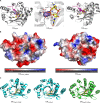
Substrate binding by EAL domains. (A) Ribbon diagram structure of the EAL domains of BlrP1, a fully functional class I PDE activated by light (44), and YahA (41) binding to the substrate cyclic di-GMP. In the middle is an enlarged view of the cyclic di-GMP binding site of BlrP1. Cations are shown in violet and pink. Cyclic di-GMP is shown as sticks with carbon atoms colored yellow. (B) Comparison of electrostatic surface representations of class III EAL domains FimX of P. aeruginosa and YdiV of E. coli. While the cyclic di-GMP binding site of class IIIa FimX is conserved (model is shown with cyclic di-GMP bound), the cyclic di-GMP binding pocket is not conserved in class IIIb member YdiV. The electrostatic surface potential shows highly electronegative (red) and electropositive (blue) patches of the two proteins. (C) Ribbon diagram structure of three class IIIa cyclic di-GMP binding EAL domains: EALFimX_PSEAE (
Q9HUK6
of P. aeruginosa), EALXcFimX (
A0A0H2X6E4
of Xanthomonas campestris pv. campestris), and EALLapD_Psfluor (
Q3KK31
of Pseudomonas fluorescens Pf0-1). Note the different conformations and binding modes of cyclic di-GMP, which is displayed as sticks with carbon atoms in yellow, oxygen in red, phosphate in orange, and nitrogen in blue.
FIG 6
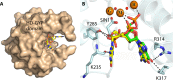
Substrate binding by the HD-GYP domain of _Pm_GH. (A) Surface representation of the _Pm_GH HD-GYP domain monomer subunit showing the binding cavity for cyclic di-GMP, which is represented in stick mode and colored by atom type. (B) Superposition of the structures of _Pm_GH bound to cyclic di-GMP and GMP. Both nucleotides are shown in stick mode. Bonding interactions are represented by dashed lines. The central metal iron has been labeled as the middle site (M) and the two flanking metal sites have been labeled H and G to reflect their proximity to the HD and GYP motifs, respectively. Residues that interact with cyclic di-GMP include Y285 of the GYP motif. Red spheres represent solvent and SIN-1, a succinate molecule (71).
FIG 7
Primary sequence alignment of HD-GYP domains from proteins that have been characterized structurally and/or enzymatically reveals the diversity within the domain. Protein designations are given in the supplemental material. The top line indicates the helices in the structure of _Pm_GH, with an annotation of the interhelix loops. Metal ligands are given in red, and proposed catalytic residues are given in green. The GYP motif is highlighted in cyan and the substrate binding ligands are in magenta. The region of the sequences with consensus motifs E(D)TG/YTY is highlighted in yellow. Note that these are not fully conserved. The triangle points to the E residue in PA4781 that may act in steric hindrance of cyclic di-GMP binding.
Similar articles
- More than Enzymes That Make or Break Cyclic Di-GMP-Local Signaling in the Interactome of GGDEF/EAL Domain Proteins of Escherichia coli.
Sarenko O, Klauck G, Wilke FM, Pfiffer V, Richter AM, Herbst S, Kaever V, Hengge R. Sarenko O, et al. mBio. 2017 Oct 10;8(5):e01639-17. doi: 10.1128/mBio.01639-17. mBio. 2017. PMID: 29018125 Free PMC article. - The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases.
Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. Lovering AL, et al. mBio. 2011 Oct 11;2(5):e00163-11. doi: 10.1128/mBio.00163-11. Print 2011. mBio. 2011. PMID: 21990613 Free PMC article. - Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP.
Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Christen M, et al. J Biol Chem. 2005 Sep 2;280(35):30829-37. doi: 10.1074/jbc.M504429200. Epub 2005 Jul 1. J Biol Chem. 2005. PMID: 15994307 - The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants.
Dow JM, Fouhy Y, Lucey JF, Ryan RP. Dow JM, et al. Mol Plant Microbe Interact. 2006 Dec;19(12):1378-84. doi: 10.1094/MPMI-19-1378. Mol Plant Microbe Interact. 2006. PMID: 17153922 Review. - Cyclic di-GMP signalling and the regulation of bacterial virulence.
Ryan RP. Ryan RP. Microbiology (Reading). 2013 Jul;159(Pt 7):1286-1297. doi: 10.1099/mic.0.068189-0. Epub 2013 May 23. Microbiology (Reading). 2013. PMID: 23704785 Free PMC article. Review.
Cited by
- Asymmetric activation mechanism of a homodimeric red light-regulated photoreceptor.
Gourinchas G, Heintz U, Winkler A. Gourinchas G, et al. Elife. 2018 Jun 5;7:e34815. doi: 10.7554/eLife.34815. Elife. 2018. PMID: 29869984 Free PMC article. - Evolution of cyclic di-GMP signalling on a short and long term time scale.
Römling U, Cao LY, Bai FW. Römling U, et al. Microbiology (Reading). 2023 Jun;169(6):001354. doi: 10.1099/mic.0.001354. Microbiology (Reading). 2023. PMID: 37384391 Free PMC article. - An emerging role for cyclic dinucleotide phosphodiesterase and nanoRNase activities in Mycoplasma bovis: Securing survival in cell culture.
Zhu X, Baranowski E, Dong Y, Li X, Hao Z, Zhao G, Zhang H, Lu D, A Rasheed M, Chen Y, Hu C, Chen H, Sagné E, Citti C, Guo A. Zhu X, et al. PLoS Pathog. 2020 Jun 29;16(6):e1008661. doi: 10.1371/journal.ppat.1008661. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32598377 Free PMC article. - Draft Genome Sequence of the Urinary Catheter Isolate Enterobacter ludwigii CEB04 with High Biofilm Forming Capacity.
Shafeeq S, Wang X, Lünsdorf H, Brauner A, Römling U. Shafeeq S, et al. Microorganisms. 2020 Apr 5;8(4):522. doi: 10.3390/microorganisms8040522. Microorganisms. 2020. PMID: 32260576 Free PMC article. - Expression and function of the cdgD gene, encoding a CHASE-PAS-DGC-EAL domain protein, in Azospirillum brasilense.
Cruz-Pérez JF, Lara-Oueilhe R, Marcos-Jiménez C, Cuatlayotl-Olarte R, Xiqui-Vázquez ML, Reyes-Carmona SR, Baca BE, Ramírez-Mata A. Cruz-Pérez JF, et al. Sci Rep. 2021 Jan 12;11(1):520. doi: 10.1038/s41598-020-80125-3. Sci Rep. 2021. PMID: 33436847 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources