Understanding development and stem cells using single cell-based analyses of gene expression - PubMed (original) (raw)
Review
Understanding development and stem cells using single cell-based analyses of gene expression
Pavithra Kumar et al. Development. 2017.
Abstract
In recent years, genome-wide profiling approaches have begun to uncover the molecular programs that drive developmental processes. In particular, technical advances that enable genome-wide profiling of thousands of individual cells have provided the tantalizing prospect of cataloging cell type diversity and developmental dynamics in a quantitative and comprehensive manner. Here, we review how single-cell RNA sequencing has provided key insights into mammalian developmental and stem cell biology, emphasizing the analytical approaches that are specific to studying gene expression in single cells.
Keywords: Computational biology; Gene regulatory networks; Pseudotime; RNA-Seq; Single cell; Stem cells.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
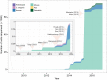
Fig. 1.
The growth of single cell genome-wide profiling techniques. A surge in scRNA-Seq applications can be observed. The cumulative number of cells that have been subjected to scRNA-Seq is shown, separated by species. Landmark studies are highlighted. Tang et al. (2009), Tang (2010b), Islam (2011), Ramskold (2012) and Hashimshony (2012) are the first five scRNA-Seq studies. They introduced the major varieties of scRNA-Seq: Tang protocol, STRT-Seq, CEL-Seq and Smart-Seq. Yan (2013) and Xue (2013) leverage scRNA-Seq to explore and the dynamics of human zygotic genome activation. Picelli (2013) introduces Smart-Seq2 with increased sensitivity. Macosko (2015) and Klein (2015) introduce high-throughput low-cost droplet-based methods that have vastly increased the number of cells that can be sequenced.
Fig. 2.
Typical approaches for analyzing scRNA-Seq datasets. Several types of analyses are popular for analyzing scRNA-Seq datasets. (A) When trying to identify cell types, dimension reduction techniques such as independent component analysis, principal component analysis, t-distributed stochastic neighbor embedding, ZIFA (Pierson and Yau, 2015) or weighted gene co-expression network analysis (Langfelder and Horvath, 2008) are first used to project high-dimensional data into a smaller number of dimensions to ease visual evaluation and interpretation. Clusters of similar cells can be identified using generally applicable methods, such as Gaussian mixture modeling (Fraley and Raftery, 2002) or K-means clustering, or methods devised specifically for single cell data, such as StemID (Grün et al., 2016), SCUBA, SNN-Cliq (Xu and Su, 2015), Destiny (Angerer et al., 2015) or BackSpin (Zeisel et al., 2015). Clusters can then be annotated based on domain-specific knowledge of the expression of a few genes, or automatically based on gene set enrichment. Finally, specific genes that are differentially expressed between clusters can be identified using scRNA-Seq-specific methods such as SCDE (Kharchenko et al., 2014) and MAST (Finak et al., 2015). (B) Most pseudotime analyses (which place each cell on a statistically derived axis that represents progression along a process, such as developmental time) start by performing dimension reduction. They then determine trajectories through the reduced dimensionality data; some algorithms identify bifurcation points and generate a distinct trajectory. The trajectories can then be used to order single cells along the process and to identify candidate regulators of stage transitions, for example, by finding stage-specific transcription factors (TF1-TF5). (C) One of the major drawbacks of scRNA-Seq is the loss of spatial context information when cells are dissociated and/or isolated. Spatial reconstruction methods attempt to ameliorate this issue by leveraging prior knowledge of landmark gene expression. Typically, localized expression of select genes is generated from in situ hybridization. Spatial reconstruction algorithms then compare scRNA-Seq profiles to discretized in situ hybridization profiles, and cells are placed in silico in the anatomical region with a matching profile. Machine-learning approaches can be used to estimate the expression of landmark genes to overcome the noisy nature of scRNA-Ssq data.
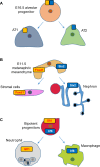
Fig. 3.
The ‘Janus’ progenitor state. scRNA-Seq has enabled the identification of embryonic progenitors that simultaneously express genes that were previously suspected of being lineage specific. (A) The PCA analysis of scRNA-Seq profiles of 198 developing murine lung cells has identified a cluster that expresses markers for both AT1 and AT2 cells, corroborating with single-cell qPCR data of E16.5 alveolar progenitors. (B) scRNA-Seq profiles of E11.5 metanephric mesenchyme has identified cells that co-express Foxd1 and Six2, which mark stromal-committing cells and nephron-committing cells, respectively. (C) A binary cell fate decision between the macrophage lineage and the neutrophil lineage was unveiled when bipotent progenitors were shown to co-express Irf8 and Gfi1, which regulate macrophage and neutrophil specification, respectively.
Similar articles
- LEAP: constructing gene co-expression networks for single-cell RNA-sequencing data using pseudotime ordering.
Specht AT, Li J. Specht AT, et al. Bioinformatics. 2017 Mar 1;33(5):764-766. doi: 10.1093/bioinformatics/btw729. Bioinformatics. 2017. PMID: 27993778 Free PMC article. - Single-cell technologies sharpen up mammalian stem cell research.
Hoppe PS, Coutu DL, Schroeder T. Hoppe PS, et al. Nat Cell Biol. 2014 Oct;16(10):919-27. doi: 10.1038/ncb3042. Nat Cell Biol. 2014. PMID: 25271480 Review. - Lineage Inference and Stem Cell Identity Prediction Using Single-Cell RNA-Sequencing Data.
Sagar, Grün D. Sagar, et al. Methods Mol Biol. 2019;1975:277-301. doi: 10.1007/978-1-4939-9224-9_13. Methods Mol Biol. 2019. PMID: 31062315 - Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations.
van den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, Robin C, van Oudenaarden A. van den Brink SC, et al. Nat Methods. 2017 Sep 29;14(10):935-936. doi: 10.1038/nmeth.4437. Nat Methods. 2017. PMID: 28960196 No abstract available. - Genomic approaches to early embryogenesis and stem cell biology.
Aiba K, Carter MG, Matoba R, Ko MS. Aiba K, et al. Semin Reprod Med. 2006 Nov;24(5):330-9. doi: 10.1055/s-2006-952155. Semin Reprod Med. 2006. PMID: 17123228 Review.
Cited by
- A single-cell atlas of the miracidium larva of Schistosoma mansoni reveals cell types, developmental pathways, and tissue architecture.
Attenborough T, Rawlinson KA, Diaz Soria CL, Ambridge K, Sankaranarayanan G, Graham J, Cotton JA, Doyle SR, Rinaldi G, Berriman M. Attenborough T, et al. Elife. 2024 Aug 27;13:RP95628. doi: 10.7554/eLife.95628. Elife. 2024. PMID: 39190022 Free PMC article. - GRouNdGAN: GRN-guided simulation of single-cell RNA-seq data using causal generative adversarial networks.
Zinati Y, Takiddeen A, Emad A. Zinati Y, et al. Nat Commun. 2024 May 14;15(1):4055. doi: 10.1038/s41467-024-48516-6. Nat Commun. 2024. PMID: 38744843 Free PMC article. - Gene Editing in Hematopoietic Stem Cells.
Liao J, Wu Y. Liao J, et al. Adv Exp Med Biol. 2023;1442:177-199. doi: 10.1007/978-981-99-7471-9_11. Adv Exp Med Biol. 2023. PMID: 38228965 Review. - Self-supervised deep clustering of single-cell RNA-seq data to hierarchically detect rare cell populations.
Lei T, Chen R, Zhang S, Chen Y. Lei T, et al. Brief Bioinform. 2023 Sep 22;24(6):bbad335. doi: 10.1093/bib/bbad335. Brief Bioinform. 2023. PMID: 37769630 Free PMC article. - Integration of a multi-omics stem cell differentiation dataset using a dynamical model.
van den Berg PR, Bérenger-Currias NMLP, Budnik B, Slavov N, Semrau S. van den Berg PR, et al. PLoS Genet. 2023 May 11;19(5):e1010744. doi: 10.1371/journal.pgen.1010744. eCollection 2023 May. PLoS Genet. 2023. PMID: 37167320 Free PMC article.
References
- Bandura D. R., Baranov V. I., Ornatsky O. I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J. E. and Tanner S. D. (2009). Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 81, 6813-6822. 10.1021/ac901049w - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical