Yersinia pestis halotolerance illuminates plague reservoirs - PubMed (original) (raw)
Yersinia pestis halotolerance illuminates plague reservoirs
Maliya Alia Malek et al. Sci Rep. 2017.
Abstract
The plague agent Yersinia pestis persists for years in the soil. Two millennia after swiping over Europe and North Africa, plague established permanent foci in North Africa but not in neighboring Europe. Mapping human plague foci reported in North Africa for 70 years indicated a significant location at <3 kilometers from the Mediterranean seashore or the edge of salted lakes named chotts. In Algeria, culturing 352 environmental specimens naturally containing 0.5 to 70 g/L NaCl yielded one Y. pestis Orientalis biotype isolate in a 40 g/L NaCl chott soil specimen. Core genome SNP analysis placed this isolate within the Y. pestis branch 1, Orientalis biovar. Culturing Y. pestis in broth steadily enriched in NaCl indicated survival up to 150 g/L NaCl as L-form variants exhibiting a distinctive matrix assisted laser desorption-ionization time-of-flight mass spectrometry peptide profile. Further transcriptomic analyses found the upregulation of several outer-membrane proteins including TolC efflux pump and OmpF porin implied in osmotic pressure regulation. Salt tolerance of Y. pestis L-form may play a role in the maintenance of natural plague foci in North Africa and beyond, as these geographical correlations could be extended to 31 plague foci in the northern hemisphere (from 15°N to 50°N).
Figures
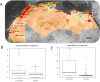
Figure 1
(A) Location of human plague foci in six countries in North Africa, 1940–2015. Plague foci are significantly located <3 km of salt source (Mediterranean sea and chotts). This figure was generated from the map of the software Google Maps/Google Earth and Google Maps/Google Earth APIs (
https://www.google.com/permissions/geoguidelines.html
).  Salt water,
Salt water,  Fresh water;
Fresh water;  1–10 cases;
1–10 cases;  10–100 cases;
10–100 cases;  >100 cases. (B) Boxplot of minimum distances to plague foci (y axis, distance in km). (C) Boxplot of mean distances to plague foci (y axis, distance in km).
>100 cases. (B) Boxplot of minimum distances to plague foci (y axis, distance in km). (C) Boxplot of mean distances to plague foci (y axis, distance in km).
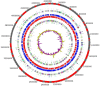
Figure 2. Circular genome map of Y. pestis Algeria3.
From outside to center: Contigs (red/grey), COGs category of genes on forward strand (three circles), genes on forward strand (blue circle), genes on reverse strand (red circle), COGs category on reverse strand (three circles), G + C content. COGs, Clusters of Orthologous Groups database.
Figure 3. Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins.
Figure 4. Phylogenetic tree of 134 Y. pestis genomes based on the 2,298 SNPs.
Biovar were designated as follows: ORI (Orientalis), ANT (Antiqua), In (intermediate strains between ANT and ORI), MED (Medievalis) and PE (Pestoides). Each strain was referred as branch 1, 2, 3 or 4 according to Cui et al..
Figure 5
Yersinia pestis forms filamentous colonies in 150 g/L NaCl-broth (A) and small colonies (B): left panel, control; right panel, Y. pestis exposed to 150 g/L NaCl.
Figure 6
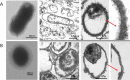
Electron microscopy of Y. pestis control (A) and Y. pestis exposed for seven weeks to 150 g/L NaCl (B). Red arrows point to enlarged cell wall, thicker in control cells (A) than in Y. pestis cells exposed to 150 g/L NaCl (B).
Figure 7
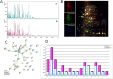
Proteomics of Y. pestis exposed to 150 g/L NaCl: (A): MALDI-TOF mass spectrometry (a: control; b: exposed) (B). Representative 2D differential gel electrophoresis (DIGE) analysis of Y. pestis proteins (C): string network of DIGE analysis of Y. pestis proteins. Each individual sample from Y. pestis Orientalis wild type and 150 g/L NaCl exposed Y. pestis and a pooled reference sample were labeled with Cy5, Cy3, and Cy2, respectively, and were then separated on the same gel using the 2D-DIGE system. Three images were obtained from each gel and an overlay of dye scan images was also obtained. Selected protein spots exhibiting an ANOVA score lower or close to 0.05 and a change of at least 1.5-fold intensity are indicated by circles and spot numbers as indicated in Table (D): Analysis, according the COG family, of upregulated and downregulated proteins identified by mass spectrometry after 2D-differential gel electrophoresis separation.
Similar articles
- Long-term persistence of virulent Yersinia pestis in soil.
Ayyadurai S, Houhamdi L, Lepidi H, Nappez C, Raoult D, Drancourt M. Ayyadurai S, et al. Microbiology (Reading). 2008 Sep;154(Pt 9):2865-2871. doi: 10.1099/mic.0.2007/016154-0. Microbiology (Reading). 2008. PMID: 18757820 - Persistence of Yersinia pestis in soil under natural conditions.
Eisen RJ, Petersen JM, Higgins CL, Wong D, Levy CE, Mead PS, Schriefer ME, Griffith KS, Gage KL, Beard CB. Eisen RJ, et al. Emerg Infect Dis. 2008 Jun;14(6):941-3. doi: 10.3201/eid1406.080029. Emerg Infect Dis. 2008. PMID: 18507908 Free PMC article. - Plague in the genomic area.
Drancourt M. Drancourt M. Clin Microbiol Infect. 2012 Mar;18(3):224-30. doi: 10.1111/j.1469-0691.2012.03774.x. Clin Microbiol Infect. 2012. PMID: 22369155 Review. - Ecology of Yersinia pestis and the Epidemiology of Plague.
Dubyanskiy VM, Yeszhanov AB. Dubyanskiy VM, et al. Adv Exp Med Biol. 2016;918:101-170. doi: 10.1007/978-94-024-0890-4_5. Adv Exp Med Biol. 2016. PMID: 27722862 Review. - Plague: A Millenary Infectious Disease Reemerging in the XXI Century.
Grácio AJDS, Grácio MAA. Grácio AJDS, et al. Biomed Res Int. 2017;2017:5696542. doi: 10.1155/2017/5696542. Epub 2017 Aug 20. Biomed Res Int. 2017. PMID: 28904964 Free PMC article. Review.
Cited by
- Different characteristics of the soil in marmot habitats might be one of the factors that influcting Yersinia pestis prevalent in which than pikas.
Zhao W, Li S, Sun Y, Liu J, Ma Y, Qi R. Zhao W, et al. Front Microbiol. 2024 Oct 18;15:1489125. doi: 10.3389/fmicb.2024.1489125. eCollection 2024. Front Microbiol. 2024. PMID: 39493844 Free PMC article. - The Natural and Clinical History of Plague: From the Ancient Pandemics to Modern Insights.
Bennasar-Figueras A. Bennasar-Figueras A. Microorganisms. 2024 Jan 11;12(1):146. doi: 10.3390/microorganisms12010146. Microorganisms. 2024. PMID: 38257973 Free PMC article. Review. - The reproduction of gram-negative protoplasts and the influence of environmental conditions on this process.
Kanaparthi D, Lampe M, Krohn JH, Zhu B, Klingl A, Lueders T. Kanaparthi D, et al. iScience. 2023 Oct 6;26(11):108149. doi: 10.1016/j.isci.2023.108149. eCollection 2023 Nov 17. iScience. 2023. PMID: 37942012 Free PMC article. - VP3 Phage Combined with High Salt Promotes the Lysis of Biofilm-Associated Vibrio cholerae.
Li X, Li X, Zhang H, Kan B, Fan F. Li X, et al. Viruses. 2023 Jul 27;15(8):1639. doi: 10.3390/v15081639. Viruses. 2023. PMID: 37631982 Free PMC article. - No evidence for persistent natural plague reservoirs in historical and modern Europe.
Stenseth NC, Tao Y, Zhang C, Bramanti B, Büntgen U, Cong X, Cui Y, Zhou H, Dawson LA, Mooney SJ, Li D, Fell HG, Cohn S, Sebbane F, Slavin P, Liang W, Tong H, Yang R, Xu L. Stenseth NC, et al. Proc Natl Acad Sci U S A. 2022 Dec 20;119(51):e2209816119. doi: 10.1073/pnas.2209816119. Epub 2022 Dec 12. Proc Natl Acad Sci U S A. 2022. PMID: 36508668 Free PMC article.
References
- Raoult D., Mouffok N., Bitam I., Piarroux R. & Drancourt M. Plague: history and contemporary analysis. J. Infect. 66, 18–26 (2013). - PubMed
- Yersin A. La peste bubonique à Hong Kong. Ann. Inst. Pasteur 2, 428–430 (1894).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical