Flavivirus Infection Uncouples Translation Suppression from Cellular Stress Responses - PubMed (original) (raw)
. 2017 Jan 10;8(1):e02150-16.
doi: 10.1128/mBio.02150-16.
Vera Magg 1, Fabian Uch 1, Pascal Mutz 1, Philipp Klein 1, Katharina Haneke 2 3, Volker Lohmann 1, Ralf Bartenschlager 1 4, Oliver T Fackler 5, Nicolas Locker 6, Georg Stoecklin 2 3, Alessia Ruggieri 7
Affiliations
- PMID: 28074025
- PMCID: PMC5225315
- DOI: 10.1128/mBio.02150-16
Flavivirus Infection Uncouples Translation Suppression from Cellular Stress Responses
Hanna Roth et al. mBio. 2017.
Erratum in
- Erratum for Roth et al., "Flavivirus Infection Uncouples Translation Suppression from Cellular Stress Responses".
Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, Stoecklin G, Ruggieri A. Roth H, et al. mBio. 2017 Apr 18;8(2):e00488-17. doi: 10.1128/mBio.00488-17. mBio. 2017. PMID: 28420740 Free PMC article. No abstract available.
Abstract
As obligate parasites, viruses strictly depend on host cell translation for the production of new progeny, yet infected cells also synthesize antiviral proteins to limit virus infection. Modulation of host cell translation therefore represents a frequent strategy by which viruses optimize their replication and spread. Here we sought to define how host cell translation is regulated during infection of human cells with dengue virus (DENV) and Zika virus (ZIKV), two positive-strand RNA flaviviruses. Polysome profiling and analysis of de novo protein synthesis revealed that flavivirus infection causes potent repression of host cell translation, while synthesis of viral proteins remains efficient. Selective repression of host cell translation was mediated by the DENV polyprotein at the level of translation initiation. In addition, DENV and ZIKV infection suppressed host cell stress responses such as the formation of stress granules and phosphorylation of the translation initiation factor eIF2α (α subunit of eukaryotic initiation factor 2). Mechanistic analyses revealed that translation repression was uncoupled from the disruption of stress granule formation and eIF2α signaling. Rather, DENV infection induced p38-Mnk1 signaling that resulted in the phosphorylation of the eukaryotic translation initiation factor eIF4E and was essential for the efficient production of virus particles. Together, these results identify the uncoupling of translation suppression from the cellular stress responses as a conserved strategy by which flaviviruses ensure efficient replication in human cells.
Importance: For efficient production of new progeny, viruses need to balance their dependency on the host cell translation machinery with potentially adverse effects of antiviral proteins produced by the infected cell. To achieve this, many viruses evolved mechanisms to manipulate host cell translation. Here we find that infection of human cells with two major human pathogens, dengue virus (DENV) and Zika virus (ZIKV), leads to the potent repression of host cell translation initiation, while the synthesis of viral protein remains unaffected. Unlike other RNA viruses, these flaviviruses concomitantly suppress host cell stress responses, thereby uncoupling translation suppression from stress granule formation. We identified that the p38-Mnk1 cascade regulating phosphorylation of eIF4E is a target of DENV infection and plays an important role in virus production. Our results define several molecular interfaces by which flaviviruses hijack host cell translation and interfere with stress responses to optimize the production of new virus particles.
Copyright © 2017 Roth et al.
Figures
FIG 1
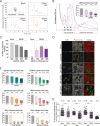
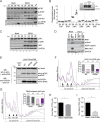
Induction of host translation repression by flavivirus infection. (A) Representative polysome profiles of naive Huh7 cells (Mock) and DENV-infected cells at 18, 24, and 36 h p.i. Cell extracts of naive Huh7 cells or cells infected with DENV (MOI of 10) for the indicated time periods were loaded on a sucrose gradient and separated by ultracentrifugation. Sucrose gradients were eluted from the top using a fractionator, and absorption at 254 nm was continuously recorded. Shown in the Mock panel is the separation between actively translated, polysomal mRNAs associated with multiple ribosomes and not or poorly translated subpolysomal mRNAs (40S and 60S, single ribosomal subunits; 80S, monosome). DENV-infected cells show an increase of the monosomal 80S peak throughout the infection. (B) Representative polysome profile analysis (lower panel). The percentage of polysomal ribosomes (actively translating mRNAs) is assessed by measuring the area below the polysomal part of the curve and the area of subpolysomal and polysomal parts of the curve. Histogram bars shown in the upper panel represent the mean percentages of polysomal ribosomes ± standard error of the mean (SEM). n, number of profiles analyzed. (C) Abundance of specific mRNAs in gradient fractions. Polysome profiles of naive (Mock) and DENV-infected Huh7 cells were recorded at the indicated time points p.i. Total RNA was extracted from all fractions, and the relative abundance of specific mRNAs in each fraction was quantified by qRT-PCR. Histogram bars represent mean percentages of GAPDH mRNA and DENV positive-strand RNA genome associated with the polysomal fractions ± standard deviation (SD) (n = 3). (D) Reduction of protein synthesis in DENV-infected cells. Naive Huh7 cells (Mock) and cells infected with DENV for the indicated time period were treated with puromycin to induce a premature release of nascent polypeptidic chains. Arsenite-treated Huh7 cells were used as a control. Puromycylated chains are visualized using an antipuromycin antibody (red) and infection by immunostaining of DENV NS3 (green). Representative fields of view are shown. Yellow squares represent the cropped section shown in the merge panel. Scale bars, 50 µm. (E) Scatter plot of de novo protein synthesis measured by fluorescence intensity of the puromycin signal (mean fluorescence intensities ± SD; n = 3) a.u., arbitrary units. Statistical significance and the number of analyzed cells (n) are given at the top. ***, P < 0.001; **, P < 0.01. (F) Host cell translation repression is a general feature of flavivirus infection. Polysomal profiles of Huh7 cells infected with DENV serotype 1, 3, or 4, WNV strain NY, or ZIKV MR766 or H/PF/2013 (MOI of 10) for the indicated time periods were recorded (see Fig. S2 in the supplemental material). Shown are mean percentages of polysomal ribosomes ± SEM. n, number of profiles analyzed.
FIG 2
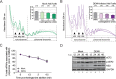
Host cell translation is impaired at the initiation step by DENV infection. (A, B, and C) DENV infection does not affect RNA translation elongation. Naive Huh7 cells (Mock [A]) and cells infected with DENV (MOI of 10) for 24 h (B) were treated with harringtonine for 1.5, 3, and 4.5 min to allow ribosome runoff. Treatment with DMSO for 4.5 min was used as a control. Shown are representative polysome profile analyses (lower panel) and mean percentages of polysomal ribosomes ± SEM (upper panel; n, number of profiles analyzed). (C) Mean percentages ± SD (n = 3) of total mRNAs associated with polysomes in mock- and DENV-infected cells upon harringtonine treatment (corresponding to panels A and B) were normalized to the values of DMSO-treated cells, respectively. (D) Phosphorylation levels of the eEF2 are not affected by DENV infection. Shown is representative Western blot analysis (n = 2) of Huh7 cells infected with DENV (MOI of 10) for 12, 24, 36, and 48 h (lanes 5 to 8). Naive Huh7 cells (Mock) cultured in parallel for the same time periods were used as reference (lanes 1 to 4).
FIG 3
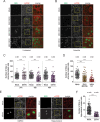
Flavivirus infection inhibits eIF2α-dependent and -independent SG formation in Huh7 cells (A, B, and C). Huh7 cells were infected with DENV (MOI of 0.5) for the indicated time period and left untreated (A) or treated with arsenite (B) before fixation. DENV infection was visualized by immunostaining of NS5 (green) and SGs by immunostaining of eIF3B (red). Naive cells (Mock) served as control. Representative fields of view are shown. Yellow squares represent the cropped section shown in the merge panel. Scale bar, 50 µm. (C) Scatter plot displaying the number of SGs in cells treated with arsenite. Shown are mean values ± SD of a representative experiment (n = 3). n, number of cells analyzed. ***, P < 0.001; n.s., not significant. (D) ZIKV and WNV inhibit arsenite-induced SG assembly. Similar to panel C, Huh7 cells were infected with ZIKV MR766 or WNV NY (MOI of 0.5) for 24 h and treated with arsenite before fixation. (E and F) Huh7 cells were infected with DENV (MOI of 0.5) for 30 h and treated with hippuristanol for 8 h before fixation. (E) DENV infection was visualized by immunostaining of NS5 (green) and SGs by immunostaining of eIF3B (red). Naive cells (Mock) served as control. Representative fields of view are shown. Yellow squares represent the cropped section shown in the merge panel. Scale bars, 50 µm. (F) Scatter plot displaying the number of SGs in cells treated with hippuristanol. Shown are mean values ± SD from a representative experiment (n = 3). n, number of cells analyzed. ***, P < 0.001.
FIG 4
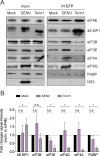
DENV-induced host cell translation repression is independent of the PKR-eIF2α signaling pathway. (A and B) activation of PKR by DENV does not result in eIF2α phosphorylation. Cells were infected with DENV (MOI of 10) for 12, 24, 36, and 48 h (lanes 5 to 8). Naive Huh7 cells (Mock) cultured in parallel for the same time periods were used as reference (lanes 1 to 4). Shown are representative Western blot analyses (n = 4). (A) Analysis of PKR and phospho-PKR (p-PKR) abundance. Cells transfected with the synthetic dsRNA poly(I-C) (lane 9) were used as a positive control. (B) Analysis of eIF2α and phospho-eIF2α (p-eIF2α) abundance. Cells treated with arsenite (lane 9) were used as a positive control. Phosphorylation of eIF2α was analyzed by Phos-tag acrylamide gel (lower panel). (C) DENV-induced translational repression is PKR independent. Polysome profiles of Huh7 PKR ko cells (clone 2#3) left untreated (Mock) or infected with DENV (MOI of 10) were recorded at the indicated times. Shown are representative polysome profile analyses (lower panel) and mean percentages of polysomal ribosomes ± SEM (upper panel). n, number of profiles analyzed. (D) DENV-induced translational repression is eIF2α independent. Polysome profiles of Huh7 control cells (Ctrl) and Huh7 cells stably expressing GADD34 and infected with DENV (MOI of 10) were recorded 24 h p.i. Naive cells (Mock) were used as a control. Shown are representative polysome profile analyses (lower panel) and mean percentages of polysomal ribosomes ± SEM (upper panel). n, number of profiles analyzed.
FIG 5
Cap-binding complex assembly is not affected by DENV infection. (A) m7GTP immunoprecipitation from naive Huh7 cells or cells infected with DENV (MOI of 10) for 24 h. Cells treated with Torin1 for 16 h were used as control of cap-binding complex disassembly. Shown is representative Western blot analysis of cap-binding proteins coimmunoprecipitated with m7GTP-immobilized agarose beads. Shown are input cell extracts (1% of total, left panel) and immunoprecipitated proteins (25% of eluate, right panel). (B) Quantification of cap-binding proteins associated with m7GTP. Shown are means ± SEM of fold changes (n = 3). *, P < 0.05; n.s., not significant.
FIG 6
Activation of the p38-Mnk1 signaling pathway is required for virus production. (A) eIF4E phosphorylation levels increase in DENV-infected cells. Shown is representative Western blot analysis (n = 3) of phospho-eIF4E (p-eIF4E) abundance in naive and DENV-infected Huh7 cells at 12, 24, 36, and 48 h p.i. Phosphorylation of eIF4E was analyzed by Phos-tag acrylamide gel (lower panel). (B) Analysis of MAPK phosphorylation levels in Huh7 cells inoculated with UV-inactivated DENV (Ctrl) or infected with DENV for 24 and 43 h by using the Proteome Profiler human phospho-MAPK array. Shown in the lower panel are mean relative pixel densities normalized to the control of two independent experiments with two measurements each. (Upper panel) Representative Western blot analysis (n = 2) of Huh7 cell extracts used for analysis of MAPK phosphorylations. Naive Huh7 cells, Huh7 cells infected with DENV for 24 and 43 h, and Huh7 cells inoculated with UV-inactivated DENV for 43 h were analyzed. (C) Representative Western blot analysis of phospho-p38α (p-p38α) abundance in naive and DENV-infected Huh7 cells (n = 3). (D) Representative Western blot analysis of phospho-Mnk1 (p-Mnk1) abundance in naive and DENV-infected Huh7 cells (n = 2). Lysates of naive and murine norovirus (MNV)-infected mouse leukemic monocytes/macrophages (RAW264.7) served as a positive control (72). (E, F, and G) Phosphorylation of eIF4E is dispensable for DENV-induced repression of translation. (E) Ectopic expression of HA-tagged eIF4E in Huh7 cells. Shown is representative Western blot (WB) analysis (n = 3) of endogenous eIF4E and HA-eIF4E abundance in Huh7 HA-eIF4E, Huh7 HA-eIF4E(S209A), and Huh7 HA-eIF4E(S209D). Huh7 control (Ctrl) cells served as a control. (Upper panel) Immunoblotting using an anti-eIF4E antibody. (Lower panel) Immunoblotting using an anti-HA antibody. (F and G) DENV-induced translational repression is phospho-eIF4E independent. (F) Polysome profiles of Huh7 cells stably expressing HA-eIF4E (WT) and the mutant HA-eIF4E(S209A) infected with DENV (MOI of 10) were recorded 24 h p.i. Naive cells (Mock) were used as a control. Shown are representative polysome profile analyses (lower panel) and mean percentages of polysomal ribosomes ± SEM (upper panel) n, number of profiles analyzed. (G) Naive Huh7 cells (Mock) and cells infected with DENV (MOI of 10) for 8 h (DENV) were treated with CGP57380, an inhibitor of Mnk1 phosphorylation, for 16 h. Treatment with DMSO was used as a control. Shown are representative polysome profile analyses (lower panel) and mean percentages of polysomal ribosomes ± SEM (upper panel) n, number of profiles analyzed. (H) Inhibition of p38α reduces DENV virus production. Huh7 cells were infected with DENV (MOI of 0.1) and cotreated with DMSO or 50 µM SB203580, an inhibitor of p38α activity. Virus titers were determined by limiting dilution assay. Shown is the mean ± SD (n = 3). *, P < 0.05. (I) Inhibition of Mnk1 phosphorylation severely diminishes DENV virus production. Huh7 cells were infected with DENV (MOI of 0.1) for 8 h and subsequently treated with DMSO or 50 µM CGP57380 for 16 h. Virus titers were determined by limiting dilution assay (TCID50 per milliliter). Shown is the mean ± SD (n = 4). *, P < 0.05.
FIG 7
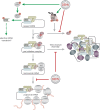
Modifications of host cell translation and stress response by DENV infection. Early in DENV infection, host cell translation is repressed. While assembly of the preinitiation complex and translation elongation remain unaffected, translation initiation is stalled after association of the 60S ribosomal subunit with cellular mRNA (monosomal RNA) (red blunted arrows). Interestingly, DENV infection activates the p38α-Mnk1 pathway, resulting in the phosphorylation of eIF4E (green arrows). While eIF4E phosphorylation does not account for DENV-induced translation repression, it might regulate the selective translation of specific mRNA subsets (green dashed arrow). DENV genome replication occurs through dsRNA intermediates that are sensed by PKR, leading to PKR autophosphorylation and activation (green arrow). However, the downstream phosphorylation of eIF2α, a direct target of the activated PKR, is inhibited as well as the assembly of both eIF2α-dependent and -independent SGs (red blunt-ended arrows). Altogether, during DENV infection translation suppression is uncoupled from the activation of the stress response.
Similar articles
- RPLP1 and RPLP2 Are Essential Flavivirus Host Factors That Promote Early Viral Protein Accumulation.
Campos RK, Wong B, Xie X, Lu YF, Shi PY, Pompon J, Garcia-Blanco MA, Bradrick SS. Campos RK, et al. J Virol. 2017 Jan 31;91(4):e01706-16. doi: 10.1128/JVI.01706-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27974556 Free PMC article. - Mouse Norovirus Infection Arrests Host Cell Translation Uncoupled from the Stress Granule-PKR-eIF2α Axis.
Fritzlar S, Aktepe TE, Chao YW, Kenney ND, McAllaster MR, Wilen CB, White PA, Mackenzie JM. Fritzlar S, et al. mBio. 2019 Jun 18;10(3):e00960-19. doi: 10.1128/mBio.00960-19. mBio. 2019. PMID: 31213553 Free PMC article. - Cotranslational prolyl hydroxylation is essential for flavivirus biogenesis.
Aviner R, Li KH, Frydman J, Andino R. Aviner R, et al. Nature. 2021 Aug;596(7873):558-564. doi: 10.1038/s41586-021-03851-2. Epub 2021 Aug 18. Nature. 2021. PMID: 34408324 Free PMC article. - Establishment and Application of Flavivirus Replicons.
Kümmerer BM. Kümmerer BM. Adv Exp Med Biol. 2018;1062:165-173. doi: 10.1007/978-981-10-8727-1_12. Adv Exp Med Biol. 2018. PMID: 29845532 Review. - AXL, an Important Host Factor for DENV and ZIKV Replication.
Xie S, Zhang H, Liang Z, Yang X, Cao R. Xie S, et al. Front Cell Infect Microbiol. 2021 Mar 22;11:575346. doi: 10.3389/fcimb.2021.575346. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33954117 Free PMC article. Review.
Cited by
- Roadblocks and fast tracks: How RNA binding proteins affect the viral RNA journey in the cell.
Girardi E, Pfeffer S, Baumert TF, Majzoub K. Girardi E, et al. Semin Cell Dev Biol. 2021 Mar;111:86-100. doi: 10.1016/j.semcdb.2020.08.006. Epub 2020 Aug 23. Semin Cell Dev Biol. 2021. PMID: 32847707 Free PMC article. Review. - Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions.
Flynn RA, Belk JA, Qi Y, Yasumoto Y, Wei J, Alfajaro MM, Shi Q, Mumbach MR, Limaye A, DeWeirdt PC, Schmitz CO, Parker KR, Woo E, Chang HY, Horvath TL, Carette JE, Bertozzi CR, Wilen CB, Satpathy AT. Flynn RA, et al. Cell. 2021 Apr 29;184(9):2394-2411.e16. doi: 10.1016/j.cell.2021.03.012. Epub 2021 Mar 11. Cell. 2021. PMID: 33743211 Free PMC article. - Stress granules: potential therapeutic targets for infectious and inflammatory diseases.
Li W, Wang Y. Li W, et al. Front Immunol. 2023 May 2;14:1145346. doi: 10.3389/fimmu.2023.1145346. eCollection 2023. Front Immunol. 2023. PMID: 37205103 Free PMC article. Review. - Zika virus inhibits eIF2α-dependent stress granule assembly.
Amorim R, Temzi A, Griffin BD, Mouland AJ. Amorim R, et al. PLoS Negl Trop Dis. 2017 Jul 17;11(7):e0005775. doi: 10.1371/journal.pntd.0005775. eCollection 2017 Jul. PLoS Negl Trop Dis. 2017. PMID: 28715409 Free PMC article. - PKR-mediated stress response enhances dengue and Zika virus replication.
Ricciardi-Jorge T, da Rocha EL, Gonzalez-Kozlova E, Rodrigues-Luiz GF, Ferguson BJ, Sweeney T, Irigoyen N, Mansur DS. Ricciardi-Jorge T, et al. mBio. 2023 Oct 31;14(5):e0093423. doi: 10.1128/mbio.00934-23. Epub 2023 Sep 21. mBio. 2023. PMID: 37732809 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials