Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis - PubMed (original) (raw)
. 2017 Mar;49(3):367-376.
doi: 10.1038/ng.3753. Epub 2017 Jan 16.
Xin Li 2, Tyler Saunders 3, Rakel Tryggvadottir 2, Samantha J Mentch 4, Marc O Warmoes 4, Anna E Word 1, Alessandro Carrer 5, Tal H Salz 2, Sonoko Natsume 1, Kimberly M Stauffer 1, Alvin Makohon-Moore 3, Yi Zhong 3, Hao Wu 6, Kathryn E Wellen 5, Jason W Locasale 4, Christine A Iacobuzio-Donahue 7, Andrew P Feinberg 2 8
Affiliations
- PMID: 28092686
- PMCID: PMC5695682
- DOI: 10.1038/ng.3753
Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis
Oliver G McDonald et al. Nat Genet. 2017 Mar.
Abstract
During the progression of pancreatic ductal adenocarcinoma (PDAC), heterogeneous subclonal populations emerge that drive primary tumor growth, regional spread, distant metastasis, and patient death. However, the genetics of metastases largely reflects that of the primary tumor in untreated patients, and PDAC driver mutations are shared by all subclones. This raises the possibility that an epigenetic process might operate during metastasis. Here we report large-scale reprogramming of chromatin modifications during the natural evolution of distant metastasis. Changes were targeted to thousands of large chromatin domains across the genome that collectively specified malignant traits, including euchromatin and large organized chromatin histone H3 lysine 9 (H3K9)-modified (LOCK) heterochromatin. Remarkably, distant metastases co-evolved a dependence on the oxidative branch of the pentose phosphate pathway (oxPPP), and oxPPP inhibition selectively reversed reprogrammed chromatin, malignant gene expression programs, and tumorigenesis. These findings suggest a model whereby linked metabolic-epigenetic programs are selected for enhanced tumorigenic fitness during the evolution of distant metastasis.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures
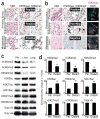
Figure 1. Global epigenetic reprogramming during the evolution of distant metastasis
(a) IHC stains against H3K9me2/3 performed on tumor sections as indicated for patient A124 (regional/peritoneal spread), which corresponds to samples detailed in Supplementary Table 1 (founder: A124PrF; subclone: A124PrS; peritoneal metastasis: A124Per). Staining was diffusely positive (brown nuclei) across all tumor sections. (b) IHC and IF stains on tumor sections from patient A125 (distant metastatic spread), which corresponds to samples detailed in Supplementary Table 1 (founder: A125PrF; subclone: A125PrS; liver metastasis: A125Lv1). There was progressive loss of nuclear staining that initiated in primary tumor subclones that seeded metastases (middle panels) with diffuse loss in the liver metastases (scale bars=100μm for IHC, 20μm for IF; IF colors: red=H3K9me2; blue=DAPI; green=vimentin). (c) Western blots on cells lines collected from a peritoneal subclone (Per) and distant (liver, lung) metastatic subclones from same patient (pA38, see Supplementary Fig. 1c for corresponding IHC) also showed reduced levels of heterochromatin modifications with increased acetylation in distant metastatic subclones compared to peritoneal. (d) Densitometry summary of western blot findings for the indicated histone modifications across cell lines from distant metastatic subclones compared to peritoneal carcinomatosis (derived from blots shown in Supplementary Fig. 1e–f, n=8 biological replicates, error bars: s.e.m., *p<0.01 by two-tailed t-tests).
Figure 2. Epigenomic reprogramming of chromatin domains during PDAC subclonal evolution
(a) Representative (left panels) and total summarized (right panels) ChIP-seq experiments revealed loss of H3K9me2 from LOCKs between peritoneal (A38Per) and distant metastatic and primary tumor precursor subclones (others). H3K27me3 remained strong in all subclones. (b) WGBS data on cell lines (A38, A13, top panel) and frozen tissue samples (A124, A125 bottom panels) showed that samples from local regional spread and parental clones (A38Per, A124PrF, A124Per, A125PrF) possessed hypermethylated LOCKs. In contrast, distant metastatic subclones (A125Lv1/2, A13Lg, A38Lv, A38Lg) and their primary tumor subclones (A125PrS, A13Pr1, A13Pr2) showed hypomethylation of DNA across the same LOCK regions. (c) Examples of individual LOCKs displaying hyper- vs. hypomethylation across subclones, as described above. WGBS signals are plotted above (0.5–1.0) and below (0.0–0.5) a midpoint of 0.5 in IGV, which represents the background (basal) level of methylation across all samples. (d) Global levels of H3K36me3, H3K27ac, and DNA methylation within ECDs did not show any consistent differences between subclones. (e) In contrast, distant metastatic subclones and primary tumor subclones (red lines) displayed local reprogramming of H3K36me3 and H3K27ac specifically over DE genes within ECDs, compared to the same DE genic ECD regions from A38Per controls (black lines) compared against each subclone independently.
Figure 3. Reprogrammed chromatin domains encode divergent malignant properties
(a) A38Lg was remarkably resistant to H202 treatments compared to A38Per. MTT signals reflect cell viability normalized to untreated controls (n=4 technical replicates, error bars: s.d.m., *p<0.03 by two tailed t-tests). (b) Western blots for proteins involved in epithelial and EMT differentiation were differentially expressed between A38Per and A38Lg, as predicted by GO analyses of reprogrammed DE genes from LOCKs. (c) A38Lg was highly resistant to gemcitabine compared to A38Per, as predicted by GO analyses of reprogrammed DE genes from ECDs. MTT signals reflect cell viability normalized to untreated controls (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). (d) A38Lg possessed elevated levels of γH2AX by western blot, consistent with activation of DNA repair pathways. (e) Western blots showed that A38Lg lost hyper-phosphorylated ERK and (f) was resistant to ERK targeted therapy, compared to A38Per. MTT signals reflect cell viability, normalized to untreated controls (n=4 technical replicates, error bars: s.d.m., *p<0.03 by two tailed t-tests). (g) A38Lg also lost sensitivity to KRAS knockdown by matrigel 3D tumor forming assays, compared to A38Per (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm.
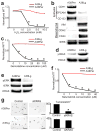
Figure 4. Hyperactive glucose metabolism and 6PG depletion in distant metastatic subclones
(a) MTT assays performed on equal numbers of viable, growth-arrested cells from the indicated subclones showed elevated signal (oxidoreductase activity) across distant metastatic subclones, compared to HPDE and regional PDACs (n=4 technical replicates, error bars: s.d.m., *p<10−5 by two tailed t-tests). (b) Normalized cell counts for the indicated samples incubated with (+) or without (−) 10mM glucose and treated with 1mM H202 as indicated (+,−) showed that HPDE cells were sensitive to H202 under either glucose condition (as expected), whereas regional PDAC samples were resistant to H202 irrespective of glucose availability (n=3 technical replicates, error bars: s.d.m). (c) In contrast, distant metastatic subclones were sensitive to H202 when glucose was not present in the media (n=3 technical replicates, error bars: s.d.m, *p<0.001 by two tailed t-tests). (d) Simplified schematic of 13C-(1,2)-labeled glucose flow through glycolysis and the PPP. Glucose that enters the oxidative branch of the PPP has one labeled carbon cleaved during conversion of 6PG to Ru5P (m+1), whereas glucose that travels through glycolysis retains both labeled carbons (m+2). Note that cross-talk allows glucose with either labeling pattern to re-enter the other pathway and incorporate. (e) LC-MS for nucleotides and lactate showed that these downstream metabolites acquired greatly elevated 13C-1,2 labels from glucose in A38Lg (n=3 biological replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). (f) Steady state LC-HRMS measurements for 6PG showed either complete (ND: not detected) or near complete loss of metabolite across distant metastases and their precursors.
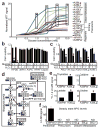
Figure 5. PGD-dependence in distant metastatic subclones
(a) Western blots against indicated histone modifications performed on peritoneal (A38Per) and distant metastatic (A38Lg) subclones from the same patient showed that global levels of reprogrammed H3K9me2 and acetylation in A38Lg were reversed by removal of glucose from the media (left panel), PGD RNAi (middle panel), and 6AN treatments (right panel). (b) Western blots on A38Lg indicated that PGD knockdown by RNAi did not perturb expression of other PPP components or KRAS. (c) PGD RNAi did not affect the ability of HPDE cells or regional PDAC samples to form tumors in 3D matrigel assays (representative photomicrographs shown with quantified numbers of tumors/well, n=4 technical replicates, error bars: s.d.m.). (d) In contrast, PGD RNAi significantly reduced tumor formation across distant metastatic subclones (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm.
Figure 6. Reversal of reprogrammed chromatin and tumorigenic potential by 6AN
(a) Densitometry summary of western blots shown in Supplementary Fig. 12a (n=8 biological replicates, error bars: s.e.m., *p≤0.01 by two tailed t-tests). 6AN selectively reversed reprogrammed H3K9me2 and H3K9/27ac. (b) Densitometry summary of western blots shown in Supplementary Fig. 12b (n=6 biological replicates, error bars: s.e.m.). 6AN had minimal effects on histone modifications across normal (HPDE, fibroblast) or regional PDAC samples. (c) 6AN ablated tumor formation in 3D matrigel assays (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm. (d) 6AN also blocked the ability of distant metastatic subclones to form tumors when injected into 3D organotypic stromal cultures (n=3 technical replicates, error bars: s.d.m., *p<0.05 by two tailed t-tests). Scale bars: 200μm.
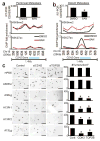
Figure 7. CDH2 and TOP2B are epigenetic targets of 6AN that selectively regulate tumorigenic potential
(a) Real-time RT-qPCR (top panel) and ChIP-qPCR for H3K9me2 and H3K27ac showed that 6AN had minimal effect on CDH2 mRNA expression or epigenetic state (H3K9me2/H3K27ac) over the CDH2 locus in the peritoneal subclone. Numbers along the x-axis of ChIP panels correspond to coordinates of the ChIP-PCR primer positions across the 1.4Mb CDH2 locus (Chr18q 27Mb:primer position). (b) In contrast, RT-PCR (top panel) showed that the distant metastatic subclone (A38Lg) from the same patient over-expressed CDH2 relative to A38Per (compare y-axis values), and that expression was repressed by 6AN (n=4 technical PCR replicates from two biological replicate experiments, error bars: s.d.m., *p=0.002 by two tailed t-tests). ChIP assays further showed that 6AN induced enrichment of H3K9me2 across the 1.4Mb CDH2 locus with corresponding reductions of H3K27ac directly over the genic region (n=2 biological replicates, error bars: s.e.m.). (c) RNAi against both CDH2 and TOP2B (Supplementary Fig. 15c–d) selectively blocked 3D tumor formation in distant metastatic and precursor subclones that over-expressed these genes by RNA-seq, with no effect on A38Per or HPDE cells (n=4 technical replicates, error bars: s.d.m., *p<0.01 by two tailed t-tests). Scale bars: 400μm.
Comment in
- Tumour evolution: Epigenetic and genetic heterogeneity in metastasis.
Alderton GK. Alderton GK. Nat Rev Cancer. 2017 Feb 23;17(3):141. doi: 10.1038/nrc.2017.11. Nat Rev Cancer. 2017. PMID: 28228644 No abstract available. - Pancreatic Cancer Genomics 2.0: Profiling Metastases.
Collisson EA, Maitra A. Collisson EA, et al. Cancer Cell. 2017 Mar 13;31(3):309-310. doi: 10.1016/j.ccell.2017.02.014. Cancer Cell. 2017. PMID: 28292434
Similar articles
- Pentose conversions support the tumorigenesis of pancreatic cancer distant metastases.
Bechard ME, Word AE, Tran AV, Liu X, Locasale JW, McDonald OG. Bechard ME, et al. Oncogene. 2018 Sep;37(38):5248-5256. doi: 10.1038/s41388-018-0346-5. Epub 2018 May 30. Oncogene. 2018. PMID: 29849117 Free PMC article. - Epigenomic Changes Prime PDAC Metastasis.
[No authors listed] [No authors listed] Cancer Discov. 2017 Apr;7(4):OF3. doi: 10.1158/2159-8290.CD-NB2017-025. Epub 2017 Feb 23. Cancer Discov. 2017. PMID: 28232366 - Epigenetic Alterations in Pancreatic Cancer Metastasis.
Wang SS, Xu J, Ji KY, Hwang CI. Wang SS, et al. Biomolecules. 2021 Jul 22;11(8):1082. doi: 10.3390/biom11081082. Biomolecules. 2021. PMID: 34439749 Free PMC article. Review. - Epigenetic Landscape in Pancreatic Ductal Adenocarcinoma: On the Way to Overcoming Drug Resistance?
Ciernikova S, Earl J, García Bermejo ML, Stevurkova V, Carrato A, Smolkova B. Ciernikova S, et al. Int J Mol Sci. 2020 Jun 8;21(11):4091. doi: 10.3390/ijms21114091. Int J Mol Sci. 2020. PMID: 32521716 Free PMC article. Review. - Evolution and dynamics of pancreatic cancer progression.
Yachida S, Iacobuzio-Donahue CA. Yachida S, et al. Oncogene. 2013 Nov 7;32(45):5253-60. doi: 10.1038/onc.2013.29. Epub 2013 Feb 18. Oncogene. 2013. PMID: 23416985 Free PMC article. Review.
Cited by
- Global landscape of 2-hydroxyisobutyrylation in human pancreatic cancer.
Lu Y, Li X, Zhao K, Qiu P, Deng Z, Yao W, Wang J. Lu Y, et al. Front Oncol. 2022 Sep 30;12:1001807. doi: 10.3389/fonc.2022.1001807. eCollection 2022. Front Oncol. 2022. PMID: 36249039 Free PMC article. - Epigenomic heterogeneity as a source of tumour evolution.
Laisné M, Lupien M, Vallot C. Laisné M, et al. Nat Rev Cancer. 2025 Jan;25(1):7-26. doi: 10.1038/s41568-024-00757-9. Epub 2024 Oct 16. Nat Rev Cancer. 2025. PMID: 39414948 Review. - Pancreatic Cancer: Pathogenesis, Screening, Diagnosis, and Treatment.
Wood LD, Canto MI, Jaffee EM, Simeone DM. Wood LD, et al. Gastroenterology. 2022 Aug;163(2):386-402.e1. doi: 10.1053/j.gastro.2022.03.056. Epub 2022 Apr 7. Gastroenterology. 2022. PMID: 35398344 Free PMC article. Review. - H3K9me3 represses G6PD expression to suppress the pentose phosphate pathway and ROS production to promote human mesothelioma growth.
Lu C, Yang D, Klement JD, Colson YL, Oberlies NH, Pearce CJ, Colby AH, Grinstaff MW, Liu Z, Shi H, Ding HF, Liu K. Lu C, et al. Oncogene. 2022 Apr;41(18):2651-2662. doi: 10.1038/s41388-022-02283-0. Epub 2022 Mar 30. Oncogene. 2022. PMID: 35351997 Free PMC article. - Emerging epigenomic landscapes of pancreatic cancer in the era of precision medicine.
Lomberk G, Dusetti N, Iovanna J, Urrutia R. Lomberk G, et al. Nat Commun. 2019 Aug 28;10(1):3875. doi: 10.1038/s41467-019-11812-7. Nat Commun. 2019. PMID: 31462645 Free PMC article. Review.
References
- Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. - PubMed
METHODS REFERENCES
- Li C, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227.e5. - PubMed
MeSH terms
Substances
Grants and funding
- F31 CA180682/CA/NCI NIH HHS/United States
- R01 CA179991/CA/NCI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R00 CA168997/CA/NCI NIH HHS/United States
- R01 CA054358/CA/NCI NIH HHS/United States
- P50 CA095103/CA/NCI NIH HHS/United States
- R01 CA140599/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases