Metabolomics and Metabolic Diseases: Where Do We Stand? - PubMed (original) (raw)
Review
Metabolomics and Metabolic Diseases: Where Do We Stand?
Christopher B Newgard. Cell Metab. 2017.
Abstract
Metabolomics, or the comprehensive profiling of small molecule metabolites in cells, tissues, or whole organisms, has undergone a rapid technological evolution in the past two decades. These advances have led to the application of metabolomics to defining predictive biomarkers for incident cardiometabolic diseases and, increasingly, as a blueprint for understanding those diseases' pathophysiologic mechanisms. Progress in this area and challenges for the future are reviewed here.
Keywords: cardiovascular diseases; diabetes; metabolic diseases; metabolism; metabolomics; obesity.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
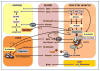
Figure 1. Emergent mechanisms of branched-chain amino acid (BCAA) metabolism in cardiometabolic disease pathogenesis unveiled with metabolomics
Several mechanisms contribute to accumulation of BCAA in plasma of obese, insulin resistant humans, including increased de novo production of BCAA by the gut microbiome and reduced utilization of BCAA in liver and adipose tissue. BCAA utilization does not appear to be suppressed in skeletal muscle, and under obese conditions, elevated BCAA induce a decrease in skeletal muscle glycine levels, removing a potential escape valve for excess acyl CoAs, Combined substrate pressure from elevated BCAA and lipids in obesity contribute to accumulation of incompletely oxidized fatty acids in mitochondria (“mitochondrial overload”) and reduced efficiency of glucose disposal. In addition, valine catabolism yields two new BCAA-derived factors that contribute to energy balance and metabolic homeostasis--β-aminoisobutyric acid (BAIBA), which stimulates thermogenesis and browning of white fat, and 3-hydroxyisobutryate (3-HIB), which stimulates trans-endothelial and muscle uptake of fatty acids. See text for details and discussion.
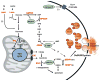
Figure 2. Emergent mechanisms of glucose-stimulated insulin secretion (GSIS) from metabolomics studies
Application of metabolomics methods to the pancreatic islet has resulted in identification of mechanisms that may complement the classical KATP-channel-dependent pathway for GSIS. This includes a pathway initiated by anaplerotic metabolism of glucose-derived pyruvate through pyruvate carboxylase (PC), egress of citrate, isocitrate, and a-ketoglutarate from the mitochondria to the cytosol via the citrate/isocitrate carrier (CIC), engagement of isocitrate with the cytosolic, NADP-dependent isoform of isocitrate dehydrogenase (IDH1) and reduction of glutathione to GSH by glutathione reductase. A second pathway involving metabolism of glucose through the pentose monophosphate shunt, including the first two NAPDH producing steps glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH), results in a sharp increase in adenylosuccinate (S-AMP) produced from IMP via the adenylosuccinate synthase (ADSS) reaction. Importantly, intermediates generated by either the isocitrate/GSH (isocitrate, NADPH, GSH) or S-AMP (S-AMP) pathways stimulate exocytosis in permeabilized human β-cells and rescue loss of glucose regulation in β-cells from humans with type 2 diabetes. Also for both pathways, the effects on exocytosis require expression of the insulin granule desumoylating enzyme SENP1. Also shown, metabolomics has identified two metabolites associated with risk of type 2 diabetes that modulate β-cell function, 2-aminoadipic acid (2-AAA), which enhances insulin secretion at basal glucose levels, and the fatty acid furan metabolite, 3-carboxy-4-methyl-5-propyl-2- furanpropanoic acid (CMPF), which impairs glucose metabolism and insulin secretion. See text for details and discussion.
References
- Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. - PubMed
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl CoA decarboxylase reverses muscle, liver, and whole-animal insulin resistance. Nature Medicine. 2004;10:268–274. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 DK078669/DK/NIDDK NIH HHS/United States
- P01 DK058398/DK/NIDDK NIH HHS/United States
- R24 DK085610/DK/NIDDK NIH HHS/United States
- R37 DK046492/DK/NIDDK NIH HHS/United States
- R01 DK046492/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous