Cytoplasmic m6A reader YTHDF3 promotes mRNA translation - PubMed (original) (raw)
. 2017 Mar;27(3):444-447.
doi: 10.1038/cr.2017.10. Epub 2017 Jan 20.
Yu-Sheng Chen 1 2, Xiao-Li Ping 1 2, Xin Yang 1 2, Wen Xiao 1 2, Ying Yang 1 2, Hui-Ying Sun 1 2, Qin Zhu 1 2, Poonam Baidya 1 2, Xing Wang 1 2, Devi Prasad Bhattarai 1 2, Yong-Liang Zhao 3, Bao-Fa Sun 1 3, Yun-Gui Yang 1 2
Affiliations
- PMID: 28106076
- PMCID: PMC5339832
- DOI: 10.1038/cr.2017.10
Cytoplasmic m6A reader YTHDF3 promotes mRNA translation
Ang Li et al. Cell Res. 2017 Mar.
No abstract available
Figures
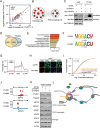
Figure 1
YTHDF3 promotes translation by interacting with ribosomal proteins. (A) Scatter plot of proteins bound to YTHDF1 versus YTHDF3. Enriched 40S and 60S subunit proteins are highlighted in blue and red, respectively. (B) Gene ontology and enrichment analysis of proteins that interact with both YTHDF1 and YTHDF3. 152 proteins were subjected to DAVID GO analysis. An enrichment map was constructed by using Cytoscape with the Enrichment Map plugin. (C) PAR-CLIP of YTHDF3 protein in control- and YTHDF1-depleted HeLa cells transfected with Myc-YTHDF3 plasmid. The pull-down RNA products were labeled with biotin at 3′ end (Biotinylation Kit, Thermo) and then visualized by the Chemiluminescent nucleic acid detection module. (D) Venn diagram of the overlapping mRNAs with binding clusters of YTHDF1 and YTHDF3 detected by PAR-CLIP-seq. (E) Gene ontology analysis of 2 945 mRNAs bound by both YTHDF1 and YTHDF3. DAVID was used for the GO analysis. The enriched terms were ranked by −log10 (_P_-value). (F) YTHDF1 and YTHDF3 binding motifs identfied by HOMER from PAR-CLIP clusters. (G) Distribution of YTHDF1 and YTHDF3 binding clusters across the length of mRNA transcripts. 5′ UTRs, CDSs and 3 UTRs of mRNAs are individually binned into regions spanning 1% of their total length, and the percentage of YTHDF1 and YTHDF3 binding clusters that fall within each bin is determined. (H) Immunofluorescence analysis of nascent protein synthesis in control- and YTHDF3-deficient HeLa cells. FITC (green): newly synthesized protein; CY3 (red): 5′-CY3-labeled siRNAs; DAPI (blue): DNA. Scale bar: 10 μm. Representative images from one of three independent experiments are shown. (I) Cumulative distribution of translation efficiency (ratio of ribosome-bound fragments and mRNA input) among non-targets of both YTHDF1 and YTHDF3, YTHDF1 unique targets, YTHDF3 unique targets and YTHDF1/3 common targets. The _p_-values are calculated using a two-sided Mann-Whitney test. (J) IGV tracks displaying YTHDF1, YTHDF3 binding clusters and m6A peaks within EEF1G and LRPAP1. The green dots at the bottom of the tracks depict the positions of m6A peaks. (K) Protein levels of YTHDF1/3 common targets (EEF1G and LRPAP1), YTHDF3 unique targets (ADAR1 and EIF2S3) and non-targets of both YTHDF1 and YTHDF3 (RPS15 and EIF4E) in YTHDF1 or YTHDF3 knockdown cells detected by western blotting. HeLa cells were transfected with YTHDF1, YTHDF3 or control siRNA. Forty eight hours later, cells were lysed and subjected to western blotting with the indicated antibodies. (L) YTHDF3, in cooperation with YTHDF1, modulates the translation of m6A-modified mRNAs by binding to m6A-modified mRNAs and interacting with proteins of 40S and 60S subunits.
Similar articles
- N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. Wang X, et al. Cell. 2015 Jun 4;161(6):1388-99. doi: 10.1016/j.cell.2015.05.014. Cell. 2015. PMID: 26046440 Free PMC article. - YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. Shi H, et al. Cell Res. 2017 Mar;27(3):315-328. doi: 10.1038/cr.2017.15. Epub 2017 Jan 20. Cell Res. 2017. PMID: 28106072 Free PMC article. - m6A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance.
Lobo J, Costa AL, Cantante M, Guimarães R, Lopes P, Antunes L, Braga I, Oliveira J, Pelizzola M, Henrique R, Jerónimo C. Lobo J, et al. J Transl Med. 2019 Mar 12;17(1):79. doi: 10.1186/s12967-019-1837-z. J Transl Med. 2019. PMID: 30866959 Free PMC article. - The role of RNA-binding and ribosomal proteins as specific RNA translation regulators in cellular differentiation and carcinogenesis.
Ceci M, Fazi F, Romano N. Ceci M, et al. Biochim Biophys Acta Mol Basis Dis. 2021 Apr 1;1867(4):166046. doi: 10.1016/j.bbadis.2020.166046. Epub 2020 Dec 28. Biochim Biophys Acta Mol Basis Dis. 2021. PMID: 33383105 Review. - Cytoplasmic fate of eukaryotic mRNA: identification and characterization of AU-binding proteins.
Jarzembowski JA, Malter JS. Jarzembowski JA, et al. Prog Mol Subcell Biol. 1997;18:141-72. doi: 10.1007/978-3-642-60471-3_7. Prog Mol Subcell Biol. 1997. PMID: 8994264 Review. No abstract available.
Cited by
- RNA Binding by the m6A Methyltransferases METTL16 and METTL3.
Mansfield KD. Mansfield KD. Biology (Basel). 2024 May 29;13(6):391. doi: 10.3390/biology13060391. Biology (Basel). 2024. PMID: 38927271 Free PMC article. Review. - N6-methyladenosine regulates PEDV replication and host gene expression.
Chen J, Jin L, Wang Z, Wang L, Chen Q, Cui Y, Liu G. Chen J, et al. Virology. 2020 Sep;548:59-72. doi: 10.1016/j.virol.2020.06.008. Epub 2020 Jun 16. Virology. 2020. PMID: 32838947 Free PMC article. - The extensive m5C epitranscriptome of Thermococcus kodakarensis is generated by a suite of RNA methyltransferases that support thermophily.
Fluke KA, Fuchs RT, Tsai YL, Talbott V, Elkins L, Febvre HP, Dai N, Wolf EJ, Burkhart BW, Schiltz J, Brett Robb G, Corrêa IR Jr, Santangelo TJ. Fluke KA, et al. Nat Commun. 2024 Aug 23;15(1):7272. doi: 10.1038/s41467-024-51410-w. Nat Commun. 2024. PMID: 39179532 Free PMC article. - FMRP phosphorylation modulates neuronal translation through YTHDF1.
Zou Z, Wei J, Chen Y, Kang Y, Shi H, Yang F, Shi Z, Chen S, Zhou Y, Sepich-Poore C, Zhuang X, Zhou X, Jiang H, Wen Z, Jin P, Luo C, He C. Zou Z, et al. Mol Cell. 2023 Dec 7;83(23):4304-4317.e8. doi: 10.1016/j.molcel.2023.10.028. Epub 2023 Nov 9. Mol Cell. 2023. PMID: 37949069 Free PMC article. - The Role of m6A Modification and m6A Regulators in Esophageal Cancer.
Li Y, Niu C, Wang N, Huang X, Cao S, Cui S, Chen T, Huo X, Zhou R. Li Y, et al. Cancers (Basel). 2022 Oct 20;14(20):5139. doi: 10.3390/cancers14205139. Cancers (Basel). 2022. PMID: 36291923 Free PMC article. Review.
References
- Xiao W, Adhikari S, Dahal U, et al. Mol Cell 2016; 61:507–519. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases