InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines - PubMed (original) (raw)
InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines
Quan Li et al. Am J Hum Genet. 2017.
Abstract
In 2015, the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) published updated standards and guidelines for the clinical interpretation of sequence variants with respect to human diseases on the basis of 28 criteria. However, variability between individual interpreters can be extensive because of reasons such as the different understandings of these guidelines and the lack of standard algorithms for implementing them, yet computational tools for semi-automated variant interpretation are not available. To address these problems, we propose a suite of methods for implementing these criteria and have developed a tool called InterVar to help human reviewers interpret the clinical significance of variants. InterVar can take a pre-annotated or VCF file as input and generate automated interpretation on 18 criteria. Furthermore, we have developed a companion web server, wInterVar, to enable user-friendly variant interpretation with an automated interpretation step and a manual adjustment step. These tools are especially useful for addressing severe congenital or very early-onset developmental disorders with high penetrance. Using results from a few published sequencing studies, we demonstrate the utility of InterVar in significantly reducing the time to interpret the clinical significance of sequence variants.
Keywords: ACMG; ANNOVAR; ClinVar; InterVar; clinical interpretation; genetic diagnosis; variant annotation; variant interpretation.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
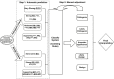
Figure 1
Flowchart of the Two-Step Procedure of InterVar Underlined and bold fonts denote automated criteria.
Figure 2
Illustration of the 28 Criteria from the 2015 ACMG-AMP Guidelines For some criteria, the name of the internal database and its size are denoted within parentheses.
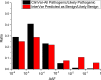
Figure 3
AAF Distribution of Pathogenic or Likely Pathogenic ClinVar Variants Predicted to Be Benign or Likely Benign by InterVar and All Pathogenic or Likely Pathogenic ClinVar Variants
Figure 4
Illustration of wInterVar (A) Automatic interpretation of genetic variants, which can be entered by several means. (B) Once users click “adjust,” the full list of criteria is shown for manual adjustment, after which the final results are given.
Similar articles
- Adapting ACMG/AMP sequence variant classification guidelines for single-gene copy number variants.
Brandt T, Sack LM, Arjona D, Tan D, Mei H, Cui H, Gao H, Bean LJH, Ankala A, Del Gaudio D, Knight Johnson A, Vincent LM, Reavey C, Lai A, Richard G, Meck JM. Brandt T, et al. Genet Med. 2020 Feb;22(2):336-344. doi: 10.1038/s41436-019-0655-2. Epub 2019 Sep 19. Genet Med. 2020. PMID: 31534211 - Clinical Interpretation of Sequence Variants.
Zhang J, Yao Y, He H, Shen J. Zhang J, et al. Curr Protoc Hum Genet. 2020 Jun;106(1):e98. doi: 10.1002/cphg.98. Curr Protoc Hum Genet. 2020. PMID: 32176464 Free PMC article. - Genebe.net: Implementation and validation of an automatic ACMG variant pathogenicity criteria assignment.
Stawiński P, Płoski R. Stawiński P, et al. Clin Genet. 2024 Aug;106(2):119-126. doi: 10.1111/cge.14516. Epub 2024 Mar 5. Clin Genet. 2024. PMID: 38440907 - The current state of clinical interpretation of sequence variants.
Hoskinson DC, Dubuc AM, Mason-Suares H. Hoskinson DC, et al. Curr Opin Genet Dev. 2017 Feb;42:33-39. doi: 10.1016/j.gde.2017.01.001. Epub 2017 Jan 31. Curr Opin Genet Dev. 2017. PMID: 28157586 Free PMC article. Review. - Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Richards S, et al. Genet Med. 2015 May;17(5):405-24. doi: 10.1038/gim.2015.30. Epub 2015 Mar 5. Genet Med. 2015. PMID: 25741868 Free PMC article.
Cited by
- Landscape of Secondary Findings in Chinese Population: A Practice of ACMG SF v3.0 List.
Huang Y, Liu B, Shi J, Zhao S, Xu K, Sun L, Chen N, Tian W, Zhang J, Wu N. Huang Y, et al. J Pers Med. 2022 Sep 14;12(9):1503. doi: 10.3390/jpm12091503. J Pers Med. 2022. PMID: 36143288 Free PMC article. - Frequent CHD1 deletions in prostate cancers of African American men is associated with rapid disease progression.
Diossy M, Tisza V, Li H, Sahgal P, Zhou J, Sztupinszki Z, Young D, Nousome D, Kuo C, Jiang J, Chen Y, Ebner R, Sesterhenn IA, Moncur JT, Chesnut GT, Petrovics G, Klus GT, Valcz G, Nuzzo PV, Ribli D, Börcsök J, Prosz A, Krzystanek M, Ried T, Szuts D, Rizwan K, Kaochar S, Pathania S, D'Andrea AD, Csabai I, Srivastava S, Freedman ML, Dobi A, Spisak S, Szallasi Z. Diossy M, et al. NPJ Precis Oncol. 2024 Sep 19;8(1):208. doi: 10.1038/s41698-024-00705-8. NPJ Precis Oncol. 2024. PMID: 39294262 Free PMC article. - Identification of a novel MAP3K1 variant in a family with 46, XY DSD and partial growth hormone deficiency.
Cheng Y, Xu C, Yang J, Zhou X, Chen N. Cheng Y, et al. Mol Med Rep. 2022 Nov;26(5):338. doi: 10.3892/mmr.2022.12854. Epub 2022 Sep 14. Mol Med Rep. 2022. PMID: 36102299 Free PMC article. - GPCards: An integrated database of genotype-phenotype correlations in human genetic diseases.
Li B, Wang Z, Chen Q, Li K, Wang X, Wang Y, Zeng Q, Han Y, Lu B, Zhao Y, Zhang R, Jiang L, Pan H, Luo T, Zhang Y, Fang Z, Xiao X, Zhou X, Wang R, Zhou L, Wang Y, Yuan Z, Xia L, Guo J, Tang B, Xia K, Zhao G, Li J. Li B, et al. Comput Struct Biotechnol J. 2021 Mar 22;19:1603-1611. doi: 10.1016/j.csbj.2021.03.011. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33868597 Free PMC article. - Broadening the scope of multigene panel analysis for adult epilepsy patients.
Lee S, Kang MK, So KH, Jang R, Shin YW, Jang SS, Yoon JG, Kim S, Kim M, Chu K, Lee SK, Kim KJ, Baek ST, Lim BC, Moon J. Lee S, et al. Epilepsia Open. 2024 Aug;9(4):1538-1549. doi: 10.1002/epi4.12993. Epub 2024 Jul 1. Epilepsia Open. 2024. PMID: 38946282 Free PMC article.
References
- McPherson J.D. Next-generation gap. Nat. Methods. 2009;6(11, Suppl):S2–S5. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases