Diversity of DNA Replication in the Archaea - PubMed (original) (raw)
Review
Diversity of DNA Replication in the Archaea
Darya Ausiannikava et al. Genes (Basel). 2017.
Abstract
DNA replication is arguably the most fundamental biological process. On account of their shared evolutionary ancestry, the replication machinery found in archaea is similar to that found in eukaryotes. DNA replication is initiated at origins and is highly conserved in eukaryotes, but our limited understanding of archaea has uncovered a wide diversity of replication initiation mechanisms. Archaeal origins are sequence-based, as in bacteria, but are bound by initiator proteins that share homology with the eukaryotic origin recognition complex subunit Orc1 and helicase loader Cdc6). Unlike bacteria, archaea may have multiple origins per chromosome and multiple Orc1/Cdc6 initiator proteins. There is no consensus on how these archaeal origins are recognised- some are bound by a single Orc1/Cdc6 protein while others require a multi- Orc1/Cdc6 complex. Many archaeal genomes consist of multiple parts-the main chromosome plus several megaplasmids-and in polyploid species these parts are present in multiple copies. This poses a challenge to the regulation of DNA replication. However, one archaeal species (Haloferax volcanii) can survive without replication origins; instead, it uses homologous recombination as an alternative mechanism of initiation. This diversity in DNA replication initiation is all the more remarkable for having been discovered in only three groups of archaea where in vivo studies are possible.
Keywords: DNA replication; Haloferax; Orc1/Cdc6; Sulfolobus; archaea; replication origin.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the writing of the manuscript.
Figures
Figure 1
Current view of the archaeal phylogenetic tree. Based on [6,7]. The groups in which in vivo replication initiation studies have been undertaken are underlined.
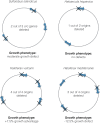
Figure 2
Binding of Orc1/Cdc6 proteins at origins of archaeal chromosomes. (A) Aeropyrum pernix Cdc6-1 binds to oriC1 as a monomer; binding to the origin oriC2 has not been investigated; (B) Cdc6-1, Cdc6-2, and Cdc6-3 of Sulfolobus solfataricus binds more than one origin each; (C) Replication initiation proteins of Sulfolobus acidocaldarius bind only one origin each. Similar to S. acidocaldarius, initiation proteins in Sulfolobus islandicus bind only one origin each.
Figure 3
Serial deletion of orc genes or origins in different archaeal species. The highest number of orc/origin deletions possible in one strain is shown. Chromosomes are not drawn to scale.
Figure 4
Recombination-dependent replication initiation. The invading 3′ DNA end of a displacement loop (D-loop) recombination intermediate is used as a primer for leading strand DNA synthesis. Formation of a D-loop requires a RecA-family recombinase.
Similar articles
- Influence of Origin Recognition Complex Proteins on the Copy Numbers of Three Chromosomes in Haloferax volcanii.
Ludt K, Soppa J. Ludt K, et al. J Bacteriol. 2018 Aug 10;200(17):e00161-18. doi: 10.1128/JB.00161-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29941422 Free PMC article. - Characterization of the replication initiator Orc1/Cdc6 from the Archaeon Picrophilus torridus.
Arora J, Goswami K, Saha S. Arora J, et al. J Bacteriol. 2014 Jan;196(2):276-86. doi: 10.1128/JB.01020-13. Epub 2013 Nov 1. J Bacteriol. 2014. PMID: 24187082 Free PMC article. - Genetic and physical mapping of DNA replication origins in Haloferax volcanii.
Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. Norais C, et al. PLoS Genet. 2007 May 18;3(5):e77. doi: 10.1371/journal.pgen.0030077. Epub 2007 Apr 5. PLoS Genet. 2007. PMID: 17511521 Free PMC article. - Archaeal orc1/cdc6 proteins.
Bell SD. Bell SD. Subcell Biochem. 2012;62:59-69. doi: 10.1007/978-94-007-4572-8_4. Subcell Biochem. 2012. PMID: 22918580 Review. - Archaeal cell cycle progress.
Lundgren M, Bernander R. Lundgren M, et al. Curr Opin Microbiol. 2005 Dec;8(6):662-8. doi: 10.1016/j.mib.2005.10.008. Epub 2005 Oct 24. Curr Opin Microbiol. 2005. PMID: 16249118 Review.
Cited by
- Intein splicing efficiency and RadA levels can control the mode of archaeal DNA replication.
Liman GLS, Lennon CW, Mandley JL, Galyon AM, Zatopek KM, Gardner AF, Santangelo TJ. Liman GLS, et al. Sci Adv. 2024 Sep 20;10(38):eadp4995. doi: 10.1126/sciadv.adp4995. Epub 2024 Sep 18. Sci Adv. 2024. PMID: 39292776 Free PMC article. - Cooperation between two modes for DNA replication initiation in the archaeon Thermococcus barophilus.
Mc Teer L, Moalic Y, Cueff-Gauchard V, Catchpole R, Hogrel G, Lu Y, Laurent S, Hemon M, Aubé J, Leroy E, Roussel E, Oberto J, Flament D, Dulermo R. Mc Teer L, et al. mBio. 2024 Apr 10;15(4):e0320023. doi: 10.1128/mbio.03200-23. Epub 2024 Feb 29. mBio. 2024. PMID: 38421162 Free PMC article. - Where and when to start: Regulating DNA replication origin activity in eukaryotic genomes.
Lee CSK, Weiβ M, Hamperl S. Lee CSK, et al. Nucleus. 2023 Dec;14(1):2229642. doi: 10.1080/19491034.2023.2229642. Nucleus. 2023. PMID: 37469113 Free PMC article. Review. - Interplay between chromosomal architecture and termination of DNA replication in bacteria.
Goodall DJ, Warecka D, Hawkins M, Rudolph CJ. Goodall DJ, et al. Front Microbiol. 2023 Jun 26;14:1180848. doi: 10.3389/fmicb.2023.1180848. eCollection 2023. Front Microbiol. 2023. PMID: 37434703 Free PMC article. Review. - Putative nucleotide-based second messengers in archaea.
van der Does C, Braun F, Ren H, Albers SV. van der Does C, et al. Microlife. 2023 Jun 5;4:uqad027. doi: 10.1093/femsml/uqad027. eCollection 2023. Microlife. 2023. PMID: 37305433 Free PMC article. Review.
References
- Aves S.J., Liu Y., Richards T.A. Evolutionary diversification of eukaryotic DNA replication machinery. Subcell. Biochem. 2012;62:19–35. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources