Advancing Top-down Analysis of the Human Proteome Using a Benchtop Quadrupole-Orbitrap Mass Spectrometer - PubMed (original) (raw)
Advancing Top-down Analysis of the Human Proteome Using a Benchtop Quadrupole-Orbitrap Mass Spectrometer
Luca Fornelli et al. J Proteome Res. 2017.
Abstract
Over the past decade, developments in high resolution mass spectrometry have enabled the high throughput analysis of intact proteins from complex proteomes, leading to the identification of thousands of proteoforms. Several previous reports on top-down proteomics (TDP) relied on hybrid ion trap-Fourier transform mass spectrometers combined with data-dependent acquisition strategies. To further reduce TDP to practice, we use a quadrupole-Orbitrap instrument coupled with software for proteoform-dependent data acquisition to identify and characterize nearly 2000 proteoforms at a 1% false discovery rate from human fibroblasts. By combining a 3 m/z isolation window with short transients to improve specificity and signal-to-noise for proteoforms >30 kDa, we demonstrate improving proteome coverage by capturing 439 proteoforms in the 30-60 kDa range. Three different data acquisition strategies were compared and resulted in the identification of many proteoforms not observed in replicate data-dependent experiments. Notably, the data set is reported with updated metrics and tools including a new viewer and assignment of permanent proteoform record identifiers for inclusion of highly characterized proteoforms (i.e., those with C-scores >40) in a repository curated by the Consortium for Top-Down Proteomics.
Keywords: AUTOPILOT; Orbitrap; data-dependent acquisition; false-discovery rate; gas-phase fractionation; mass spectrometry; medium/high; proteoform; quadrupole; top-down proteomics.
Conflict of interest statement
Notes
The authors declare the following competing financial interest(s): The authors declare a conflict and several are involved in software commercialization. Thermo Fisher Scientific is an Industrial Collaborator of the NRTDP.
All RAW data files, the UniProt formatted text file used for generating the proteoform database, and the three .tdReport files associated with this study are available at http://massive.ucsd.edu/ with the identifier MSV000079913.
Figures
Figure 1
Data acquisition strategies for top-down analysis of human proteins below 60 kDa. (A) Traditional data-dependent high/high experiments as well as medium/high experiments start with a broadband MS1 scan for the determination of precursors to be fragmented in a data-dependent top-2 fashion. Similarly, the standard version of AUTOPILOT (AP), employed as first technical replicate in the high/high study, uses by default a MS1-MS2 scheme. (B) The second and third technical replicates of the AUTOPILOT experiment are designed as a SIM march, that is, as a series of SIM scans to investigate an overall 200 m/z window between 700 and 900 m/z. Precursors are selected from online deconvolution of SIM scans. (C) Selected precursors, both from Xcalibur data-dependent or AUTOPILOT-driven acquisition, are quadrupole isolated with a narrow isolation window of 3 m/z units. (D) Selected proteoforms are subject to HCD activation with dedicated parameters for high or low MW proteins. (E) An off-line database search associates each proteoform with a C-score and determines its identification confidence through an FDR calculation based on _q_-values. Well characterized proteoforms are indicated by a unique PFR identifier.
Figure 2
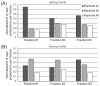
Summary of unique proteoforms and accession numbers identified at 1% FDR from 45 total RAW files. (A) Venn diagram for the total 393 unique accession numbers identified at a 1% protein-level FDR from 54 LC–MS runs. Note, ~80% of the proteins identified by medium/high experiments were not found in either of the two high/high modes of data acquisition. (B) Venn diagram of proteoforms identified at 1% FDR. Approximately 50% of identified proteoforms were shared between top-2 and AUTOPILOT high/high experiments, and low overlap was observed for the <30 kDa and 30–60 kDa portions of the fibroblast proteome interrogated here.
Figure 3
Efficiency of identification of new proteoforms from a single GELFrEE fraction using three technical replicates under Xcalibur data-dependent or AUTOPILOT data acquisition. The number of new proteoforms identified in each technical replicate for GELFrEE fractions 1, 2, and 3 is normalized over the total number of new proteoforms identified in the single GELFrEE fraction of interest. (A) The data-dependent top-2 method shows that for the three fractions considered, the first technical replicate provides the highest number of new proteoforms, and the capability of the data-dependent method of finding new confident proteoforms decreases with the number of technical replicates. (B) The AUTOPILOT experiments show that the SIM march with 50 m/z windows (2nd technical replicate) outperforms the standard AUTOPILOT acquisition based on the MS1–MS2 scheme (1st technical replicate) in two fractions out of three. Conversely, the SIM march composed by eight SIM events (3rd technical replicate) produces the lowest number of new identified proteoforms.
Figure 4
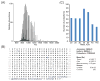
Example of ~41 kDa protein identified from a medium/high experiment (8% GELFrEE fraction 4). (A) The broadband MS1 spectrum obtained using a short transient in the Orbitrap mass analyzer shows high spectral signal-to-noise ratio for a number of charge states from 32 to 55+. (B) The graphical fragment map shows that HCD fragmentation primarily sequenced the C-terminal region to lead to a high C-score of 255 for the proteoform PFR20440, whose experimental mass matches the theoretical one within 2.5 ppm. (C) Histogram of mass distribution for proteoforms identified at 1% FDR through medium/high, top-2 experiments; the distribution is centered around 35–40 kDa.
Figure 5
C-score distributions for the three experimental setups. Identified proteoforms are binned according to their associated C-scores. Panels A–C show C-score distributions for data-dependent high–high, AUTOPILOT high/high, and data-dependent medium/high results, respectively. Proteoforms with a C-score lower than 3 are considered statistically identified but not well characterized. Proteoforms with a C-score between 3 and 40 are defined as partially characterized, as the set of fragment ions used for their identification might be consistent also with the presence of one or more highly similar proteoform(s). Finally, proteoforms with a C-scores >40 are considered well characterized, and their respective PFRs are included in a top-down proteoform repository.
Figure 6
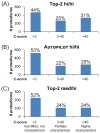
Results of Gene Ontology analysis using DAVID Bioinformatics Resources. (A) First three functional protein groups ranked according to their _p_-values. Functional groups are based on the list of UniProt accession numbers identified at 1% FDR in medium/high experiments. Note that the UniProt accession numbers of the first two functional groups are largely overlapping. (B) Mass distribution of the 41 proteoforms referring to the eight UniProt accession numbers identified for the glycolysis pathway. (C) Summary of the identified proteoforms of the glycolysis-involved enzyme L-lactate dehydrogenase (P00338).
Similar articles
- Proton Transfer Charge Reduction Enables High-Throughput Top-Down Analysis of Large Proteoforms.
Huguet R, Mullen C, Srzentić K, Greer JB, Fellers RT, Zabrouskov V, Syka JEP, Kelleher NL, Fornelli L. Huguet R, et al. Anal Chem. 2019 Dec 17;91(24):15732-15739. doi: 10.1021/acs.analchem.9b03925. Epub 2019 Nov 22. Anal Chem. 2019. PMID: 31714757 Free PMC article. - Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa.
Durbin KR, Fornelli L, Fellers RT, Doubleday PF, Narita M, Kelleher NL. Durbin KR, et al. J Proteome Res. 2016 Mar 4;15(3):976-82. doi: 10.1021/acs.jproteome.5b00997. Epub 2016 Feb 11. J Proteome Res. 2016. PMID: 26795204 Free PMC article. - Expanding Proteoform Identifications in Top-Down Proteomic Analyses by Constructing Proteoform Families.
Schaffer LV, Shortreed MR, Cesnik AJ, Frey BL, Solntsev SK, Scalf M, Smith LM. Schaffer LV, et al. Anal Chem. 2018 Jan 16;90(2):1325-1333. doi: 10.1021/acs.analchem.7b04221. Epub 2017 Dec 22. Anal Chem. 2018. PMID: 29227670 Free PMC article. - Top-Down Mass Spectrometry: Proteomics to Proteoforms.
Patrie SM. Patrie SM. Adv Exp Med Biol. 2016;919:171-200. doi: 10.1007/978-3-319-41448-5_8. Adv Exp Med Biol. 2016. PMID: 27975217 Review. - Developing top down proteomics to maximize proteome and sequence coverage from cells and tissues.
Ahlf DR, Thomas PM, Kelleher NL. Ahlf DR, et al. Curr Opin Chem Biol. 2013 Oct;17(5):787-94. doi: 10.1016/j.cbpa.2013.07.028. Epub 2013 Aug 27. Curr Opin Chem Biol. 2013. PMID: 23988518 Free PMC article. Review.
Cited by
- Online μSEC2-nRPLC-MS for Improved Sensitivity of Intact Protein Detection of IEF-Separated Nonhuman Primate Cerebrospinal Fluid Proteins.
Cline EN, Alvarez C, Duan J, Patrie SM. Cline EN, et al. Anal Chem. 2021 Dec 21;93(50):16741-16750. doi: 10.1021/acs.analchem.1c00396. Epub 2021 Dec 9. Anal Chem. 2021. PMID: 34881887 Free PMC article. - How paired PSII-LHCII supercomplexes mediate the stacking of plant thylakoid membranes unveiled by structural mass-spectrometry.
Albanese P, Tamara S, Saracco G, Scheltema RA, Pagliano C. Albanese P, et al. Nat Commun. 2020 Mar 13;11(1):1361. doi: 10.1038/s41467-020-15184-1. Nat Commun. 2020. PMID: 32170184 Free PMC article. - Single-Shot Top-Down Proteomics with Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry for Identification of Nearly 600 Escherichia coli Proteoforms.
Lubeckyj RA, McCool EN, Shen X, Kou Q, Liu X, Sun L. Lubeckyj RA, et al. Anal Chem. 2017 Nov 21;89(22):12059-12067. doi: 10.1021/acs.analchem.7b02532. Epub 2017 Nov 7. Anal Chem. 2017. PMID: 29064224 Free PMC article. - Phosphoproteomic mapping reveals distinct signaling actions and activation of muscle protein synthesis by Isthmin-1.
Zhao M, Banhos Danneskiold-Samsøe N, Ulicna L, Nguyen Q, Voilquin L, Lee DE, White JP, Jiang Z, Cuthbert N, Paramasivam S, Bielczyk-Maczynska E, Van Rechem C, Svensson KJ. Zhao M, et al. Elife. 2022 Sep 28;11:e80014. doi: 10.7554/eLife.80014. Elife. 2022. PMID: 36169399 Free PMC article. - Capillary Zone Electrophoresis-Electron-Capture Collision-Induced Dissociation on a Quadrupole Time-of-Flight Mass Spectrometer for Top-Down Characterization of Intact Proteins.
Shen X, Xu T, Hakkila B, Hare M, Wang Q, Wang Q, Beckman JS, Sun L. Shen X, et al. J Am Soc Mass Spectrom. 2021 Jun 2;32(6):1361-1369. doi: 10.1021/jasms.0c00484. Epub 2021 Mar 22. J Am Soc Mass Spectrom. 2021. PMID: 33749270 Free PMC article.
References
- Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous