Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii - PubMed (original) (raw)
Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii
Bruno P Lima et al. Microbiologyopen. 2017 Jun.
Abstract
To successfully colonize the oral cavity, bacteria must directly or indirectly adhere to available oral surfaces. Fusobacterium nucleatum plays an important role in oral biofilm community development due to its broad adherence abilities, serving as a bridge between members of the oral biofilm that cannot directly bind to each other. In our efforts to characterize the molecular mechanisms utilized by F. nucleatum to physically bind to key members of the oral community, we investigated the involvement of F. nucleatum outer membrane proteins in its ability to bind to the pioneer biofilm colonizer, Streptococcus gordonii. Here, we present evidence that in addition to the previously characterized fusobacterial adhesin RadD, the interaction between F. nucleatum ATCC 23726 and S. gordonii V288 involves a second outer membrane protein, which we named coaggregation mediating protein A (CmpA). We also characterized the role of CmpA in dual-species biofilm formation with S. gordonii V288, evaluated growth-phase-dependent as well as biofilm expression profiles of radD and cmpA, and confirmed an important role for CmpA, especially under biofilm growth conditions. Our findings underscore the complex set of specific interactions involved in physical binding and thus community integration of interacting bacterial species. This complex set of interactions could have critical implications for the formation and maturation of the oral biofilms in vivo, and could provide clues to the mechanism behind the distribution of organisms inside the human oral cavity.
Keywords: Fusobacterium nucleatum; Streptococcus gordonii; Adhesin; interspecies interaction; oral biofilm.
© 2017 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures
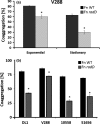
Figure 1
Quantitative coaggregation of (a) wild‐type (
WT
) Streptococcus gordonii strain V288 and the
WT
Fusobacterium nucleatum strain
ATCC
23726, or the radD mutant derivative, at different phases of growth. (b) Quantitative coaggregation of WT S. gordonii strains (
DL
1, V288,
ATCC
10558, and
ATCC
- with
WT
F. nucleatum strain
ATCC
23726, or the radD mutant derivative at exponential growth. Data represent means and standard deviation of percent coaggregation of at least three independent experiments. *p < .05 compared to wild‐type control
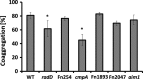
Figure 2
Quantitative coaggregation of wild‐type (
WT
) Streptococcus gordonii strain V288 with Fusobacterium nucleatum
ATCC
23726
WT
strain and
OMP
mutant derivatives (radD, Fn254, cmpA, Fn1893, Fn2047, and aim1). Data represent means and standard deviation of percent coaggregation of at least three independent experiments. *p < .05 compared to wild‐type control
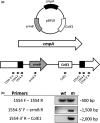
Figure 3
(a) Fusobacterium nucleatum radD cmpA double mutant (strain
BL
- was constructed by insertion of the inactivation plasmid
pBPL
9 into cmpA (Fn1554) in a radD::catP mutant background. Black arrows indicate the location of primers used for mutant construction and analysis. (b) Confirmation of plasmid insertion into cmpA by
PCR
analysis of the cmpA mutant (m) with wild‐type (WT) as the control
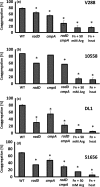
Figure 4
Quantitative coaggregation of wild‐type Streptococcus gordonii strain (a) V288, (b)
ATCC
10558, (c)
DL
1, and (d)
ATCC
51656 with Fusobacterium nucleatum strains: Wild‐type (
WT
) and the mutant derivatives: radD, cmpA, and radD cmpA double mutant. Data represent means and standard deviation of percent coaggregation of at least three independent experiments. *p < .05 compared to wild‐type control
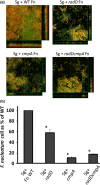
Figure 5
Streptococcus gordonii and Fusobacterium nucleatum dual‐species biofilm: (a) Confocal laser scanning microscopy of syto9‐stained dual‐species biofilm after overnight incubation. S. gordonii (Sg) cells constitutively express mCherry from their chromosome and appear orange/yellow on the images. Wild‐type (
WT
) F. nucleatum (Fn) and its mutant derivatives (radD, cmpA, and radD cmpA double mutant) are strained by syto9‐only and appear green on the images. (b) The presence of the Fn mutant strains in the Sg‐Fn dual‐species biofilm is displayed as the percentage of Fn cells normalized to the number of attached Sg cells/well, compared to that measured with
WT
Fn. Cellular ratios were determined by measuring
DNA
concentration by
qPCR
, targeting the F. nucleatum gene fomA and the S. gordonii gene srtA. At least three independent experiments were performed per strain combination. Each value represents means and standard deviation of at least three independent experiments. *p < .05 compared to wild‐type control
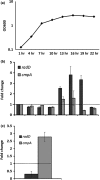
Figure 6
radD and cmpA expression:
WT
Fusobacterium nucleatum was grown in Columbia broth for 25 hr. Cell samples were collected every 3 hr and (a)
OD
600 was measured, as well as (b) radD and cmpA expression by
qRT
‐
PCR
. Gene expression was normalized to rpoB and compare to the first time point. The dashed line was added to aid in the comparison. (c) radD and cmpA expression were also measured in cells from Fn biofilm grown overnight in
SHI
‐
FSMS
. Gene expression was normalized to rpoB and compare to planktonic cells grown under the same conditions. Each value represents means and standard deviation of at least three independent experiments. To aid with visualization, the dashed line represents no change
Similar articles
- Streptococcus mutans SpaP binds to RadD of Fusobacterium nucleatum ssp. polymorphum.
Guo L, Shokeen B, He X, Shi W, Lux R. Guo L, et al. Mol Oral Microbiol. 2017 Oct;32(5):355-364. doi: 10.1111/omi.12177. Epub 2017 Feb 13. Mol Oral Microbiol. 2017. PMID: 27976528 Free PMC article. - Transcriptional responses of Streptococcus gordonii and Fusobacterium nucleatum to coaggregation.
Mutha NVR, Mohammed WK, Krasnogor N, Tan GYA, Choo SW, Jakubovics NS. Mutha NVR, et al. Mol Oral Microbiol. 2018 Dec;33(6):450-464. doi: 10.1111/omi.12248. Mol Oral Microbiol. 2018. PMID: 30329223 - The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm.
Kaplan CW, Lux R, Haake SK, Shi W. Kaplan CW, et al. Mol Microbiol. 2009 Jan;71(1):35-47. doi: 10.1111/j.1365-2958.2008.06503.x. Epub 2008 Nov 7. Mol Microbiol. 2009. PMID: 19007407 Free PMC article. - Mixed oral biofilms are controlled by the interspecies interactions of Fusobacterium nucleatum.
Valadbeigi H, Khoshnood S, Negahdari B, Maleki A, Mohammadinejat M, Haddadi MH. Valadbeigi H, et al. Oral Dis. 2024 Sep;30(6):3582-3590. doi: 10.1111/odi.14822. Epub 2023 Nov 27. Oral Dis. 2024. PMID: 38009960 Review. - Halitosis vaccines targeting FomA, a biofilm-bridging protein of fusobacteria nucleatum.
Liu PF, Huang IF, Shu CW, Huang CM. Liu PF, et al. Curr Mol Med. 2013 Sep;13(8):1358-67. doi: 10.2174/15665240113139990063. Curr Mol Med. 2013. PMID: 23865430 Review.
Cited by
- Identification of New Factors Modulating Adhesion Abilities of the Pioneer Commensal Bacterium Streptococcus salivarius.
Couvigny B, Kulakauskas S, Pons N, Quinquis B, Abraham AL, Meylheuc T, Delorme C, Renault P, Briandet R, Lapaque N, Guédon E. Couvigny B, et al. Front Microbiol. 2018 Feb 20;9:273. doi: 10.3389/fmicb.2018.00273. eCollection 2018. Front Microbiol. 2018. PMID: 29515553 Free PMC article. - The Regulatory Effect of Coaggregation Between Fusobacterium nucleatum and Streptococcus gordonii on the Synergistic Virulence to Human Gingival Epithelial Cells.
Yang R, Liu T, Pang C, Cai Y, Lin Z, Guo L, Wei X. Yang R, et al. Front Cell Infect Microbiol. 2022 Apr 29;12:879423. doi: 10.3389/fcimb.2022.879423. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35573793 Free PMC article. - Akkermansia muciniphila inhibited the periodontitis caused by Fusobacterium nucleatum.
Song B, Xian W, Sun Y, Gou L, Guo Q, Zhou X, Ren B, Cheng L. Song B, et al. NPJ Biofilms Microbiomes. 2023 Jul 17;9(1):49. doi: 10.1038/s41522-023-00417-0. NPJ Biofilms Microbiomes. 2023. PMID: 37460552 Free PMC article. - Biophysical determinants of biofilm formation in the gut.
Arias SL, Brito IL. Arias SL, et al. Curr Opin Biomed Eng. 2021 Jun;18:100275. doi: 10.1016/j.cobme.2021.100275. Epub 2021 Feb 13. Curr Opin Biomed Eng. 2021. PMID: 34504987 Free PMC article. - Streptococcus gordonii Type I Lipoteichoic Acid Contributes to Surface Protein Biogenesis.
Lima BP, Kho K, Nairn BL, Davies JR, Svensäter G, Chen R, Steffes A, Vreeman GW, Meredith TC, Herzberg MC. Lima BP, et al. mSphere. 2019 Dec 4;4(6):e00814-19. doi: 10.1128/mSphere.00814-19. mSphere. 2019. PMID: 31801844 Free PMC article.
References
- Cook, G. S. , Costerton, J. W. , & Lamont, R. J. (1998). Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii . Journal of Periodontal Research, 33, 323–327. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources