Things Are Getting Hairy: Enterobacteria Bacteriophage vB_PcaM_CBB - PubMed (original) (raw)
doi: 10.3389/fmicb.2017.00044. eCollection 2017.
Hanne Hendrix 2, Hugo Oliveira 3, Aidan Casey 4, Horst Neve 5, Olivia McAuliffe 4, R Paul Ross 4, Colin Hill 6, Jean-Paul Noben 7, Jim O'Mahony 1, Rob Lavigne 2, Aidan Coffey 1
Affiliations
- PMID: 28174560
- PMCID: PMC5259590
- DOI: 10.3389/fmicb.2017.00044
Things Are Getting Hairy: Enterobacteria Bacteriophage vB_PcaM_CBB
Colin Buttimer et al. Front Microbiol. 2017.
Abstract
Enterobacteria phage vB_PcaM_CBB is a "jumbo" phage belonging to the family Myoviridae. It possesses highly atypical whisker-like structures along the length of its contractile tail. It has a broad host range with the capability of infecting species of the genera Erwinia, Pectobacterium, and Cronobacter. With a genome of 355,922 bp, excluding a predicted terminal repeat of 22,456 bp, phage CBB is the third largest phage sequenced to date. Its genome was predicted to encode 554 ORFs with 33 tRNAs. Based on prediction and proteome analysis of the virions, 29% of its predicted ORFs could be functionally assigned. Protein comparison shows that CBB shares between 33-38% of its proteins with Cronobacter phage GAP32, coliphages PBECO4 and 121Q as well as Klebsiella phage vB_KleM_Rak2. This work presents a detailed and comparative analysis of vB_PcaM_CBB of a highly atypical jumbo myoviridae phage, contributing to a better understanding of phage diversity and biology.
Keywords: Jumbo bacteriophage; PFGE analysis; bacteriophages; bioinformatics; genome; host range; mass spectrometry; transmission electron microscopy.
Figures
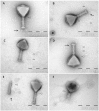
Figure 1
Electron micrographs of phage CBB with black arrows indicating baseplate fibers, open arrows indicating hair-like appendages (whiskers) and triangles indicating neck passage structure. (A,B,E) CBB virion base plate tail fibers are indicated. (B–D) Fully intact virions with atypical whisker-like structures on the contractile tail surface. (E) CBB virion contractile tail missing a capsid. (F) CBB virion with contracted tail.
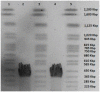
Figure 2
PFGE of phage CBB genomic DNA; lanes 1, 3, and 5 yeast chromosome PFG marker (Bio-Rad Laboratories) and lane 2 and 4 genomic DNA of phage CBB.
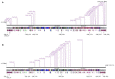
Figure 3
Comparison of the genomes of phage CBB (without predicted terminal repeat region) to other potential Rak2-like phages (Cronobacter sakazakii phage vB_CsaM_ GAP32, Escherichia coli phage 121Q, E. coli phage PBECO4, Klebsiella phage vB_KleM_RAK2, Klebsiella phage K64-1) using currently available annotations employing TBLASTX and visualised with Easyfig (Sullivan et al., 2011). A bar chart shows the G+C skew of the CBB genome, genome maps comprise of orange arrows indicating locations of genes among the different phage genomes; and lines between genome maps indicate level of homology (blue/turquoise—genes sharing orientation, red/orange—genes in inverted orientation). To assist in the comparison between genomes the largest gene of each of the phages was positioned as the first gene for each genome.
Figure 4
Maximum likelihood tree created from the alignment of the conserved region of the portal vertex region of 100 homologous sequences from different T4-like phages to that of the portal vertex protein of phage CBB, found using a BLASTP search.
Figure 5
Genome maps of bacteriophage CBB showing locations of RpoD-like promoters (A) and CBB divergent RpoD-like promoters (B) created using SnapGene.
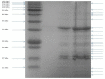
Figure 6
SDS PAGE of the structural proteins of phage vB_PcaM_CBB. Left lane shows migration patterns of New England Biolab's coloured molecular mass board range protein standard with right lanes showing that of structural proteins of phage CBB.
Similar articles
- Klebsiella phage vB_KleM-RaK2 - a giant singleton virus of the family Myoviridae.
Simoliūnas E, Kaliniene L, Truncaitė L, Zajančkauskaitė A, Staniulis J, Kaupinis A, Ger M, Valius M, Meškys R. Simoliūnas E, et al. PLoS One. 2013 Apr 9;8(4):e60717. doi: 10.1371/journal.pone.0060717. Print 2013. PLoS One. 2013. PMID: 23593293 Free PMC article. - Supersize me: Cronobacter sakazakii phage GAP32.
Abbasifar R, Griffiths MW, Sabour PM, Ackermann HW, Vandersteegen K, Lavigne R, Noben JP, Alanis Villa A, Abbasifar A, Nash JH, Kropinski AM. Abbasifar R, et al. Virology. 2014 Jul;460-461:138-46. doi: 10.1016/j.virol.2014.05.003. Epub 2014 Jun 2. Virology. 2014. PMID: 25010279 - Genomic analysis of bacteriophage PBECO4 infecting Escherichia coli O157:H7.
Kim MS, Hong SS, Park K, Myung H. Kim MS, et al. Arch Virol. 2013 Nov;158(11):2399-403. doi: 10.1007/s00705-013-1718-3. Epub 2013 May 17. Arch Virol. 2013. PMID: 23680925 - Pectobacterium carotovorum Phage vB_PcaM_P7_Pc Is a New Member of the Genus Certrevirus.
Naligama KN, Halmillawewa AP. Naligama KN, et al. Microbiol Spectr. 2022 Dec 21;10(6):e0312622. doi: 10.1128/spectrum.03126-22. Epub 2022 Nov 8. Microbiol Spectr. 2022. PMID: 36346243 Free PMC article. - Erwinia amylovora phage vB_EamM_Y3 represents another lineage of hairy Myoviridae.
Buttimer C, Born Y, Lucid A, Loessner MJ, Fieseler L, Coffey A. Buttimer C, et al. Res Microbiol. 2018 Nov;169(9):505-514. doi: 10.1016/j.resmic.2018.04.006. Epub 2018 May 18. Res Microbiol. 2018. PMID: 29777834
Cited by
- Genomic characterization of three novel Basilisk-like phages infecting Bacillus anthracis.
Farlow J, Bolkvadze D, Leshkasheli L, Kusradze I, Kotorashvili A, Kotaria N, Balarjishvili N, Kvachadze L, Nikolich M, Kutateladze M. Farlow J, et al. BMC Genomics. 2018 Sep 18;19(1):685. doi: 10.1186/s12864-018-5056-4. BMC Genomics. 2018. PMID: 30227847 Free PMC article. - Insights into the Alcyoneusvirus Adsorption Complex.
Noreika A, Rutkiene R, Dumalakienė I, Vilienė R, Laurynėnas A, Povilonienė S, Skapas M, Meškys R, Kaliniene L. Noreika A, et al. Int J Mol Sci. 2023 May 26;24(11):9320. doi: 10.3390/ijms24119320. Int J Mol Sci. 2023. PMID: 37298271 Free PMC article. - Identification of Huge Phages from Wastewater Metagenomes.
Kallies R, Hu D, Abdulkadir N, Schloter M, Rocha U. Kallies R, et al. Viruses. 2023 Nov 28;15(12):2330. doi: 10.3390/v15122330. Viruses. 2023. PMID: 38140571 Free PMC article. - Multisubunit RNA Polymerases of Jumbo Bacteriophages.
Sokolova ML, Misovetc I, V Severinov K. Sokolova ML, et al. Viruses. 2020 Sep 23;12(10):1064. doi: 10.3390/v12101064. Viruses. 2020. PMID: 32977622 Free PMC article. Review. - Larger Than Life: Isolation and Genomic Characterization of a Jumbo Phage That Infects the Bacterial Plant Pathogen, Agrobacterium tumefaciens.
Attai H, Boon M, Phillips K, Noben JP, Lavigne R, Brown PJB. Attai H, et al. Front Microbiol. 2018 Aug 14;9:1861. doi: 10.3389/fmicb.2018.01861. eCollection 2018. Front Microbiol. 2018. PMID: 30154772 Free PMC article.
References
- Ackermann H. W. (2001). Frequency of morphological phage descriptions in the year 2000. Brief review. Arch. Virol. 146, 843–857. - PubMed
- Ackermann H. W., Auclair P., Basavarajappa S., Konjin H. P., Savanurmath C. (1994). Bacteriophages from Bombyx mori. Arch. Virol. 137, 185–190. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources