Biological species are universal across Life's domains - PubMed (original) (raw)
. 2017 Feb 10;9(3):491-501.
doi: 10.1093/gbe/evx026. Online ahead of print.
Affiliations
- PMID: 28186559
- PMCID: PMC5381558
- DOI: 10.1093/gbe/evx026
Biological species are universal across Life's domains
Louis-Marie Bobay et al. Genome Biol Evol. 2017.
Abstract
Delineation of species is fundamental to organizing and understanding biological diversity. The most widely applied criterion for distinguishing species is the Biological Species Concept (BSC), which defines species as groups of interbreeding individuals that remain reproductively isolated from other such groups. The BSC has broad appeal; however, many organisms, most notably asexual lineages, cannot be classified according to the BSC. Despite their exclusively asexual mode of reproduction, Bacteria and Archaea can transfer and exchange genes though homologous recombination. Here we show that barriers to homologous gene exchange define biological species in prokaryotes with the same efficacy as in sexual eukaryotes. By analyzing the impact of recombination on the polymorphisms in thousands of genome sequences, we find that over half of named bacterial species undergo continuous recombination among sequenced constituents, indicative of true biological species. However, nearly a quarter of named bacterial species show sharp discontinuities and comprise multiple biological species. These interruptions of gene flow are not a simple function of genome identity, indicating that bacterial speciation does not uniformly proceed by the gradual divergence of genome sequences. The same genomic approach based on recombinant polymorphisms retrieves known species boundaries in sexually reproducing eukaryotes. Thus, a single biological species definition based on gene flow, once thought to be limited only to sexually reproducing organisms, is applicable to all cellular lifeforms.
Figures
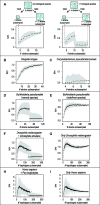
Fig. 1.—
Recognizing biological species through genome analysis. (A) “Scheme used to test for gene flow”. In each species or designated set of genomes composed of n strains (depicted as filled colored circles), nonredundant combinations of i strains (with i ranging from 4 to _n_–2 strains) were subsampled 100 times for each value of i. At each iteration, the h/m ratio was calculated for a randomly selected 10-kilobase fragment from the alignment of the core genome concatenate common to all strains. Within the bivariate plots, black dots are medians, and the grey-shaded region is the standard deviation, for the indicated number of subsampled combinations of strains. Left panel is the graphical representation observed when there are no barriers to gene flow among strains, and right panel depicts the discontinuity produced by inclusion of a strain that does not participate in gene exchange. (B–D) “Patterns of genetic exchange observed in taxonomically defined bacterial species”. Kingella kingae, in which there is gene flow among the entire set of sequenced strains; C. pseudotuberculosis, in which there is too little gene flow to assess species status; Buckholderia pseudomallei, in which there is a sharp drop in h/m ratios, denoting the presence of a sexually isolated strain or strains. The complete set of graphical representations of gene flow for 93 taxonomically defined bacterial and archaeal species is presented in supplementary fig. S1, Supplementary Material online. (E) Graphical representation of B. pseudomallei after removal of the sexually isolated strain, showing that the remaining strains constitute a biological species. The complete set of graphs is presented for the redefined species in fig. S3 before and after species redefinition. (F–I) Verification of method for recognizing biological species using obligatory sexual animals. Graphical representation of gene flow in Drosophila when including 311 haplotypes of D. melanogaster and one haplotype of D. simulans, and when the analysis is restricted to the 311 haplotypes of D. melanogaster. Graphical representation of gene flow in Homininae when including 311 haplotypes of chromosome 21 in humans and one haplotype of chimpanzees (P. troglodytes), and when the analysis is restricted to the 311 chromosome 21 haplotypes of humans.
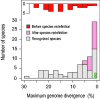
Fig. 2.—
Maximum sequence divergence within species of bacteria. Shown are the average nucleotide sequence identity values for orthologs shared by the two maximally divergent strains within each biological species of bacteria (grey and pink bars). The inset shows these values for those named species (red bars) that were subsequently redefined into biological species (pink bars) based on gene flow (n = 23). Within strictly clonal species (n = 14; not included in figure), maximally divergent strains ranged from 73 to > 99% nucleotide sequence identity (95% on average). Green dots indicate the degree of sequence identity between two maximally divergent haplotypes of D. melanogaster and of H. sapiens.
Fig. 3.—
Differential effects of sequence divergence on gene flow in biological species of bacteria. For each species, the h/m ratio for each of the subsampled combination of strains was compared to the average genome-wide sequence identity (supplementary fig. S6, Supplementary Material online). Spearman correlation coefficients Rho were computed independently for all nonredundant combinations of strains (ranging from 4 to _n_–2 strains) each subsampled 100 times and whose distributions are displayed as box-and-whiskers (first and third quartiles, 1.5 interquartile range) plots for each species. Positive values of Rho indicate a positive correlation between nucleotide sequence identity and gene flow (such that genetic exchange occurs between more similar strains), whereas negative values indicate that genetic exchange occurs preferentially among divergent strains.
Similar articles
- Life's Essential 8, Life's Simple 7 and the odds of hyperuricaemia: results from the China Multi-Ethnic Cohort Study.
Wang Y, Meng Q, Zhang X, Baima K, Chen L, Dai Y, Yang T, Feng Y, Mi F, Zhou J, Yin J. Wang Y, et al. Rheumatol Adv Pract. 2024 Jan 15;8(1):rkae009. doi: 10.1093/rap/rkae009. eCollection 2024. Rheumatol Adv Pract. 2024. PMID: 38333884 Free PMC article. - Predictive Value of Cardiovascular Health Score for Health Outcomes in Patients with PCI: Comparison between Life's Simple 7 and Life's Essential 8.
Gao X, Ma X, Lin P, Wang Y, Zhao Z, Zhang R, Yu B, Hao Y. Gao X, et al. Int J Environ Res Public Health. 2023 Feb 10;20(4):3084. doi: 10.3390/ijerph20043084. Int J Environ Res Public Health. 2023. PMID: 36833779 Free PMC article. - Life's timekeeper.
Neill D. Neill D. Ageing Res Rev. 2013 Mar;12(2):567-78. doi: 10.1016/j.arr.2013.01.004. Epub 2013 Jan 24. Ageing Res Rev. 2013. PMID: 23354279 Review. - SCALE: Skin Changes at Life's End: Final Consensus Statement: October 1, 2009.
Sibbald RG, Krasner DL, Lutz J. Sibbald RG, et al. Adv Skin Wound Care. 2010 May;23(5):225-36; quiz 237-8. doi: 10.1097/01.ASW.0000363537.75328.36. Adv Skin Wound Care. 2010. PMID: 20407297
Cited by
- High genomic differentiation and limited gene flow indicate recent cryptic speciation within the genus Laspinema (cyanobacteria).
Stanojković A, Skoupý S, Škaloud P, Dvořák P. Stanojković A, et al. Front Microbiol. 2022 Sep 9;13:977454. doi: 10.3389/fmicb.2022.977454. eCollection 2022. Front Microbiol. 2022. PMID: 36160208 Free PMC article. - Ecogenomics reveals viral communities across the Challenger Deep oceanic trench.
Zhou YL, Mara P, Vik D, Edgcomb VP, Sullivan MB, Wang Y. Zhou YL, et al. Commun Biol. 2022 Oct 4;5(1):1055. doi: 10.1038/s42003-022-04027-y. Commun Biol. 2022. PMID: 36192584 Free PMC article. - Species Identification in Plant-Associated Prokaryotes and Fungi Using DNA.
Inderbitzin P, Robbertse B, Schoch CL. Inderbitzin P, et al. Phytobiomes J. 2020;4(2):103-114. doi: 10.1094/pbiomes-12-19-0067-rvw. Epub 2020 Mar 23. Phytobiomes J. 2020. PMID: 35265781 Free PMC article. - Improved genome of Agrobacterium radiobacter type strain provides new taxonomic insight into Agrobacterium genomospecies 4.
Gan HM, Lee MVL, Savka MA. Gan HM, et al. PeerJ. 2019 Feb 8;7:e6366. doi: 10.7717/peerj.6366. eCollection 2019. PeerJ. 2019. PMID: 30775173 Free PMC article. - Biological species in the viral world.
Bobay LM, Ochman H. Bobay LM, et al. Proc Natl Acad Sci U S A. 2018 Jun 5;115(23):6040-6045. doi: 10.1073/pnas.1717593115. Epub 2018 May 21. Proc Natl Acad Sci U S A. 2018. PMID: 29784828 Free PMC article.
References
- Cain AJ. 1954. Animal species and their evolution. London: Hutchinson House.
LinkOut - more resources
Full Text Sources
Other Literature Sources