Transcriptional Addiction in Cancer - PubMed (original) (raw)
Review
Transcriptional Addiction in Cancer
James E Bradner et al. Cell. 2017.
Abstract
Cancer arises from genetic alterations that invariably lead to dysregulated transcriptional programs. These dysregulated programs can cause cancer cells to become highly dependent on certain regulators of gene expression. Here, we discuss how transcriptional control is disrupted by genetic alterations in cancer cells, why transcriptional dependencies can develop as a consequence of dysregulated programs, and how these dependencies provide opportunities for novel therapeutic interventions in cancer.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
Figure 1. Genetic changes and dysregulated gene expression programs lead to cancer cell state
The path to a cancer cell involves genetic alterations that lead to changes in the gene expression program. The dysregulated program can create dependencies on transcriptional regulators that make the tumor cells more sensitive to inhibition of these regulators than normal cells.
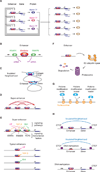
Figure 2. Key features of transcriptional regulation of gene expression programs
A) In this model of cell-type specific core regulatory circuitry, master TFs co-regulate their own genes, forming an interconnected autoregulatory loop (left), as well as those of those of many other cell-type specific genes (right). B) Enhancers are DNA elements bound by multiple TFs that recruit coactivators (such as Mediator) and RNA polymerase II, which can initiate transcription within enhancer sequences to produce enhancer RNAs (eRNAs). C) SE constituents are physically connected. Interactions among SE constituent enhancers, and between SEs and a target gene are indicated by red arcs. D) Human genome contains over 10,000 insulated neighborhoods, which are produced by multimerization of CTCF bound to two sites and reinforced with cohesin; enhancer-gene interactions occur predominantly within these neighborhoods. E) SEs are enriched for signaling transcription factors, and their associated genes, which tend to play prominent roles in cell identity, are thus especially responsive to signaling. F) Signaling TFs and other TFs that play important regulatory roles are themselves regulated by ubiquitin and proteasome-mediated destruction. G) Chromatin regulators that act through “writing”, “erasing” or “reading” histone modifications. H) DNA methylation can contribute to gene control by causing loss of TF binding at enhancers and loss of CTCF at insulated neighborhood loop anchors.
Figure 3. Components of gene control altered in cancer
Examples of three types of _cis_-factors (enhancers, promoters and insulators) and various _trans_-factors (transcription factors, cofactors, chromatin regulators, RNA polymerase II and histones) that acquire recurrent somatic mutations in cancer cells.
Figure 4. Common mechanisms of dysregulation of gene expression programs in cancer involving _trans_-factors
A) Model of a transcriptional regulatory circuit controlling the gene expression program in normal cells is shown top left. Dysregulation of gene expression programs in cancer cells can occur through dysregulation an oncogenic master TFs (bottom left), dysregulation of a transcriptional amplifier (e.g. MYC) (bottom right), and dysregulated signaling (top right). B) Model of aberrant chromatin modification affecting gene expression programs in cancer cells. C) Model of effect of cohesin mutations and CTCF mutations on enhancer-promoter interactions and insulated neighborhoods in cancer cells.
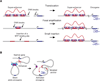
Figure 5. Common mechanisms of dysregulation of gene expression programs in cancer involving _cis_-factors
A) Mechanisms leading to the acquisition of super-enhancers to drive oncogenes in cancer cells: translocation of an existing super-enhancer, focal amplification of an enhancer element, and nucleation of super-enhancer through somatic insertion of transcription factor binding sites. B) Activation of silent proto-oncogenes by somatic mutations that disrupt insulated neighborhood anchor sites.
Figure 6. Drugging transcriptional dependencies
A) Model of the features of super-enhancer –associated oncogene control that contribute to transcriptional dependencies. B) Model of the mechanistic basis of higher transcriptional activity and vulnerability of super-enhancers: co-operative interactions between the co-activators recruited to these sites. C) Therapeutic strategy to inhibit transcriptional dependencies due to aberrant recruitment of DOT1L to driver genes in MLL-AF fusion leukemias. D) Therapeutic strategy to attack small populations of drug-tolerant tumor cells with histone deacetylase inhibitors. E) Therapeutic strategy to downregulate oncogenes through manipulation and repair of insulated neighborhoods. F) Targeted degradation of transcriptional regulators using degronimids that recruit E3-ubiquitin ligase for proteasome-mediated degradation.
Similar articles
- Molecular biology. Genetic events that shape the cancer epigenome.
Ryan RJ, Bernstein BE. Ryan RJ, et al. Science. 2012 Jun 22;336(6088):1513-4. doi: 10.1126/science.1223730. Science. 2012. PMID: 22723401 No abstract available. - Dysregulated transcription across diverse cancer types reveals the importance of RNA-binding protein in carcinogenesis.
Wang J, Liu Q, Shyr Y. Wang J, et al. BMC Genomics. 2015;16 Suppl 7(Suppl 7):S5. doi: 10.1186/1471-2164-16-S7-S5. Epub 2015 Jun 11. BMC Genomics. 2015. PMID: 26100984 Free PMC article. - Hsp90 as a "Chaperone" of the Epigenome: Insights and Opportunities for Cancer Therapy.
Isaacs JS. Isaacs JS. Adv Cancer Res. 2016;129:107-40. doi: 10.1016/bs.acr.2015.09.003. Epub 2015 Nov 24. Adv Cancer Res. 2016. PMID: 26916003 Review. - [Master and servant: epigenetic deregulations as a cause and a consequence of cancer].
Laget S, Defossez PA. Laget S, et al. Med Sci (Paris). 2008 Aug-Sep;24(8-9):725-30. doi: 10.1051/medsci/20082489725. Med Sci (Paris). 2008. PMID: 18789219 Review. French. - Single-cell analysis of clonal maintenance of transcriptional and epigenetic states in cancer cells.
Meir Z, Mukamel Z, Chomsky E, Lifshitz A, Tanay A. Meir Z, et al. Nat Genet. 2020 Jul;52(7):709-718. doi: 10.1038/s41588-020-0645-y. Epub 2020 Jun 29. Nat Genet. 2020. PMID: 32601473 Free PMC article.
Cited by
- Systematic functional interrogation of genome-wide association studies locus 17p13.3 deciphered role and genetic control of FAM57A in colorectal cancer development.
Huang J, Mo J, Xu R, Yang X, Tian Y, Ning C, Song S, Chen X, Cai Y, Zhu Y, Li B, Huang C, Jin M, Miao X. Huang J, et al. Chin J Cancer Res. 2024 Oct 30;36(5):562-576. doi: 10.21147/j.issn.1000-9604.2024.05.08. Chin J Cancer Res. 2024. PMID: 39539815 Free PMC article. - Active enhancers: recent research advances and insights into disease.
Zhang J, Wang Q, Liu J, Duan Y, Liu Z, Zhang Z, Li C. Zhang J, et al. Biol Direct. 2024 Nov 12;19(1):112. doi: 10.1186/s13062-024-00559-x. Biol Direct. 2024. PMID: 39533395 Free PMC article. Review. - FOSL1 is a key regulator of a super-enhancer driving TCOF1 expression in triple-negative breast cancer.
He Q, Hu J, Huang H, Wu T, Li W, Ramakrishnan S, Pan Y, Chan KM, Zhang L, Yang M, Wang X, Chin YR. He Q, et al. Epigenetics Chromatin. 2024 Nov 10;17(1):34. doi: 10.1186/s13072-024-00559-1. Epigenetics Chromatin. 2024. PMID: 39523372 Free PMC article. - Chromatin accessibility is associated with therapeutic response in prostate cancer.
Lee S, Lee DY, So I, Chun JN, Jeon JH. Lee S, et al. Oncol Lett. 2024 Oct 14;28(6):605. doi: 10.3892/ol.2024.14738. eCollection 2024 Dec. Oncol Lett. 2024. PMID: 39483964 Free PMC article. - Phase I First-in-Human Dose Escalation Study of the oral Casein Kinase 1α and Cyclin Dependent Kinase 7/9 inhibitor BTX-A51 in advanced MDS and AML.
Ball B, Xiao W, Borthakur G, Nguyen LXT, Valerio M, Venkatachalam A, Marcucci G, Stein A, Thai DL, Cook D, Chan K, Persaud S, Levine R, Abdel-Wahab O, Ben-Neriah Y, Stein E. Ball B, et al. Res Sq [Preprint]. 2024 Oct 15:rs.3.rs-4954060. doi: 10.21203/rs.3.rs-4954060/v1. Res Sq. 2024. PMID: 39483885 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources